Mir-21-3p and Mir-21-5p as a candidate biomarker in serum and PBMCs of patients with systemic lupus erythematosus
Naeimeh Tavakolinia , Mehdi Mahmoudi , Seyedeh Tahereh Faezi , Farshid Noorbakhsh
Affiliation
1Department of Immunology, School of Medicine, Tehran University of Medical, Tehran, Iran
2Rheumatology Research Center, Tehran University of Medical Sciences, Tehran, Iran
Corresponding Author
Maryam Izad, Professor, Department of Immunology, Ph.D-Associate School of Medicine, Tehran University of Medical Sciences, Tehran, Iran, Fax: +21-66-41-95-36, Tel: +21-64-05-32-31; E-mail: izadm@sina.tum s.ac.ir
Citation
Izad, M., et al. Mir-21-3p and Mir-21-5p as a Candidate Biomarker in Serum and PBMCs of Patients with Systemic Lupus Erythematosus (2018) J Cell Immunol Serum Biol 4(1): 4- 8.
Copy rights
© 2018 Izad, M. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Systemic lupus erythematosus; MicroRNAs; miR-21; Biomarker
Abstract
Introduction: MicroRNAs (miRNAs) are endogenous small RNAs known for their function in post-transcriptional gene regulation. miRNA regulates the differentiation and function of immune cells and its malfunction contributes to the development of various autoimmune diseases including Systemic Lupus Erythematosus (SLE). Recently, miR-21 was identified to regulate a variety of immune cells and play a crucial role in a plethora of biological functions and diseases including development, cancer and inflammation, especially correlated with the pathogenesis of autoimmune diseases such as systemic lupus erythematosus.
Material and Methods: In this report we used real-time PCR assay to compare the relative expression of mir-21-3p and mir-21-5p in SLE patients (n = 30) serums and PBMCs to healthy age and gender matched controls samples (n = 27).
Results: we found that mir-21-3p level was significantly increased in both serums and PBMCs of SLE patients compare to healthy controls (serum p value < 0.05, PBMCs p value < 0.01). But the expression level of mir-21-5p was not significantly increased in none of serums and PBMCs.
Conclusion: These data demonstrate that SLE disease is accompanied by the increased level of mir-21-3p, a subset of mir-21 and there is no relation between mir-21-5p and disease state. If these tow subsets of mir-21 have functional difference is under investigation and it’s an idea that mir-21 can be studied as a biomarker for autoimmune diseases such as SLE.
Introduction
Systemic Lupus Erythematosus (SLE) is a prototypic multisystem autoimmune disorder with a broad spectrum of clinical presentations[1] characterized by chronic immune activation and multiple immunological phenotypes[2-5]. Tissue damage is mediated by recruitment of inflammatory cells, reactive oxygen intermediates, production of inflammatory cytokines, and modulation of the coagulation cascade[6,7]. Immunologically, SLE is the consequence of loss of self-tolerance and amplification of self-antigen-mediated hyperactivation of T and B lymphocytes with more than 100 autoantibodies against numerous self-antigens including chromatin, ribonucleoproteins, and phospholipids found in patients[1]. SLE is commonly misdiagnosed as no single test is sufficiently sensitive and specific to be diagnostic or predict the beginning or end of a disease flare. Third, lack of biomarkers has impeded efforts to evaluate new SLE therapeutics in clinical trials[8]. Since the discovery of miRNAs, a growing attention has been drawn to their involvement in autoimmunity[9]. miRNAs are non-translated small single-stranded RNA molecules about 21 nucleotides in length[2,10]. That can potentially regulate every aspect of cellular activity, from differentiation and proliferation to apoptosis and function as post-transcriptional regulators of gene expression in eukaryotic organisms[11-13,3]. miRNA can directly or indirectly target a broad range of gene expression[14]. Including many immunoregulatory genes, including transcription factors, cofactors and chromatin modifiers[15,16]. through the inhibition of translation and/or decrease in mRNA stability that their changes in expression is associated with human diseases[3,11]. Due to their size, abundance, tissue specificity, and relative stability in circulation, miRNAs hold promise as unique accessible biomarkers to detect a disorder[17]. mir-21 has been classified as an oncomiR[18]. Recently, numerous investigations suggest that miR-21 plays functional roles in amounts of immune cells, such as B cells, T (Th) cells, dendritic cells (DCs)[16-19]. So dysregulation of miR-21 by several mechanisms has also been involved in various disease states, including SLE.
As the blood is easily accessible and taking a sample is lowly invasive, testable biomarkers found in blood would be especially useful and Recently, the findings that human blood contains stably expressed miRNAs[13]. One hypothesis of stability and origin of circulating miRNAs is releasing during tissue injury and the other is, miRNAs are contained in microvesicles also known as exosomes[20]. A biomarker can be defined as a genetic, biological, biochemical, or molecular event whose alterations correlate with disease pathogenesis and/or manifestations and can be evaluated qualitatively and/or quantitatively in laboratories[8]. Biomarkers may be used to facilitate accurate and early diagnosis of SLE, or identify individuals prone to develop SLE or patients at risk for severe disease and poor prognosis, some determining disease severity and / or monitoring disease progression, some may indicate systemic or specific organ involvement, and some may be used to evaluate response to treatment[8-21]. Due to the small amount of circulating miRNAs and the large amount of proteins, miRNA extraction from serum samples is technically challenging[20].
Materials and Methods
Human subjects: patients and healthy controls
We recruited 30 patients (age 37.5 ± 11.4, male/female:5 / 25): with the diagnosis of SLE who were referred to the rheumatology research center of Shariati Hospital and had been treated with the same Disease-Modifying Antirheumatic Drugs (DMARDs) or other immunosuppressive drugs. 28 healthy controls (age: 36.5 ± 13.8, male / female: 7/21) age and sex matched with no family history of SLE were also enrolled in this study. The patients were examined by specialists and informed consent was obtained from all participants for the use of their blood samples. SLEDAI for patients was equal to 1.78 ± 0.43.
Peripheral blood mononuclear cells (PBMCs) and sera were isolated from blood samples. The Hybrid-R miRNA kit (GeneALL, Korea) was used for isolation of miRNAs from 2 × 106 PBMCs according to the manufacturer’s instructions. The amount of miRNAs in serum is 106 times less than cells. Moreover, there are high amounts of proteins including enzymes, such as RNAse in serum, in this regard; we isolated miRNAs from 400 μL of serum using RNX plus (RN7713C) with modification in method. Briefly, we added 5 times more reagent (1ml per 200 μl serum) to ensure that proteins completely denaturized. Glycogen (1 μg / ul) was used as a carrier for small RNAs and as an inert reagent for the precipitation of miRNAs. The concentration and quality of total RNA were measured by a Nano Drop Spectrophotometer (Nano Drop Technologies, Waltham, US).
Total RNA (10 ng) was polyadenylated and reverse transcribed to cDNA in a final volume of 10 ul using miScript Reverse Transcription kit (Qiagen, Germanny). Realtime PCR for miR-21-3p (Qiagen,Germanny,cat.No: MS00009086) and miR-21-5p (Qiagen, Germanny,cat.No: MS00009079) was performed using miScript SYBR Green PCR kit (Qiagen, Germanny,) by the Step one PLUS instrument (ABI, US). RNU6 (Qiagen, Germanny,cat.No: MS00009086) was used as house-keeping gene. The cycle parameters for qRT-PCRs were 95°C for 15 minutes followed by 40 cycles of 94°C for 15 seconds and 55°C for 30 seconds and 70°C for 30 seconds. Expression levels of target microRNA were normalized to RNU6B gene. All primers had been bought from Qiagen.
The Ct value was calculated by subtracting the control Ct value from the SLE Ct value. The fold difference of expression between the level in control and SLE samples was calculated as 2deltaCt.
Results
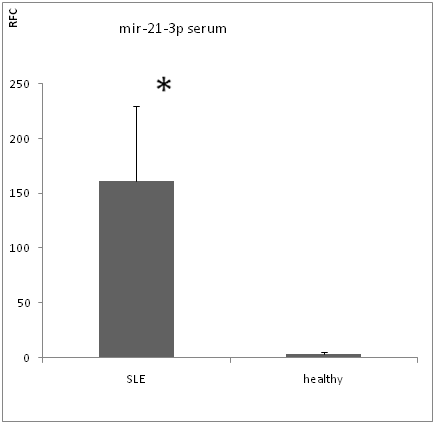
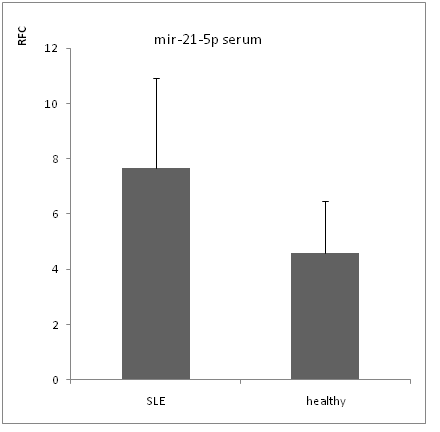
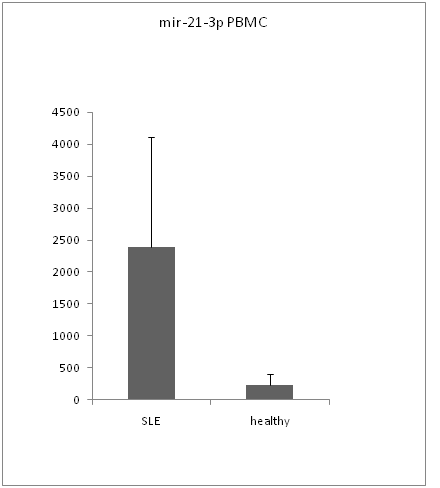
By Real-time RT-PCR analysis, we found that mir-21-3p level was significantly increased in both serums (figure 1) and PBMCs of SLE patient (figure 3) compare to healthy controls (serum p value < 0.05,PBMCs p value < 0.01). But the expression level of mir-21-5p was not significantly increased in non of serums (figure 2) and PBMCs (figure 4) with p value > 0.05.
Figure 1: mir-21-3p expression in serum.
Figure 2: mir-21-5p expression in serum.
Figure 3: mir-21-3p expression in PBMCs.
Figure 4: mir-21-5p expression in PBMCs.
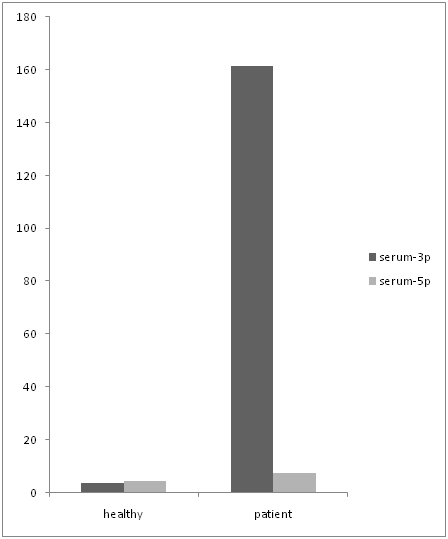
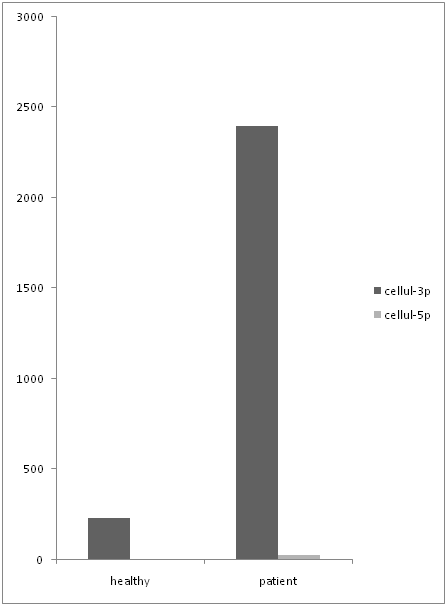
In following we had done an analysis to compare the expression level of both primers in serum and PBMC cells separately. The data shows that mir-21-3p expression was significantly higher than mir-21-5p (figure 5) in serum samples. (P Value: 0.029) even though mir-21-3p expression in PBMCs was higher than mir-21-5p (figure 6) but it doesn’t show a significant over expression. (P value > 0.05)
Figure 5: comparing the expression level of mir-21-3p and mir-21-5p in serum.
Figure 6: comparing the expression level of mir-21-3p and mir-21-5p in PBMCs.
Discussion
Expression profiling on miRNAs derived from immune cells or circulating fluid in patients with SLE likely provide useful data for understanding pathogenesis of SLE or for introducing prognostic biomarkers and novel therapeutics.
Several mechanisms of miRNAs dysregulation has also been described in different disease states, including SLE[3]. Multiple investigations show that miR-21 plays functional roles in various subsets of immune cells, such as T (Th) cells, B cells, dendritic cells (DCs)[19-21] pathogenic cytokines and immune complexes produced by Immune cells are involved in generation of autoimmune and inflammatory responses. Dysregulation of miR-21 has been also found in some autoimmune conditions[22,23]. Therefore, miR-21 is remarkably in the pathogenesis of autoimmune disorders.
PMiR-21 was also identified as the most highly expressed miRNA in human colon tissues[24,25]. Quantifying the expression of miR-21 in epidermis and dermis of psoriasis found a an increase in miR-21 expression in and dermis in comparison with healthy conrtols[26]. miR-21 expression was up-regulated in pancreatic b cells in a mouse model[23]. and miR-21 was dramatically increased in peripheral blood mononuclear cells (PBMCs) in RRMS (Relapsing Remitting Multiple Sclerosis) patients compared with controls[22] Similarly, mononuclear cells from the brains (BMNs) of Experimental Autoimmune Encephalomyelitis (EAE) mice models showed that miR-21 was increased during the acute phase of the disease and continued to be highly over expressed during the late stage of EAE[27]. Moreover, miR-21 was found higher in both the skin tissues and fibroblasts in systemic sclerosis patients[28]. miR-21 levels were dramatically increased During primary T cell activation and differentiation into Th1 and Th2 cells in vitro, and inhibition of miR-21 in T cells resulted in remarkably increase in apoptosis rate, while its overexpression reduced the apoptosis rate. Together, all these findings suggest that dysregulated expression of miR-21 may contribute to development of autoimmunity.
There are numerous findings about miR-21 involvement in SLE. Pan et al. reported that there was a much higher miR-21 expression in the splenic CD4+T cells than in the B cells isolated from MRL / lpr mice. miR-21 was increased[3]. MiR-21 in B6.Sle123 mice was constitutively increased relative controls. Furthermore its expression correlated with disease severity[19]. The other study showed that the levels of miR-21 were increased significantly in SLE patients than in healthy controls and there was more induction of miR-21 in the CD4+T in comparison to the non-CD4+T cells. MiR-21expression was positively related to disease activity[16,29]. Expression of miR-21 was higher in 5-week-old MRL/lpr mice than in 16-week-old MRL/lpr mice, when autoimmunity was established. It reveals the role miR-21 in formation of autoimmune diseases[3]. In other hand miR-21 silencing in lupus models leds to reduction of splenomegaly anh no pathologic irregularity in the spleens, livers and kidneys[19]. Stagakis et al. found over expression of PDCD4 reduced IL-10 production significantly by anti-CD3 / CD28 stimulated CD4 T cells Since PDCD4 has been shown to regulate IL-10 production, suggesting that miR-21 production in may inhibit PDCD4 in SLE patients that led to dysregulation of IL-10 production[14]. The main mechanism of PDCD4 in SLE is not clearly understood, but it is possible that PDCD4 regulates apoptotic or cell proliferation pathways contributing to the immunologic phenotype of B6.Sle123 or controls cell-signaling pathways led to hyperactive T and B cells in SLE[19].
In this study our findings about miR-21, subset 3p was closely related to the other studies that imply miR-21 significantly is overexpressed in SLE patients rather than healthy controls. Though the other subset of miR-21, mir-21-5p showed a slight increase in expression in SLE patients in comparison to controls but there was no significant evidence.
Even though the past several years have seen significant progress and tremendous success in identification and characterization of novel miRNAs critically functioning in the immune system and consequently in pathogenesis of various autoimmune diseases such as SLE, we are still in an early stage of an explosive discovery of the essential roles of these non coding small RNAs. It is noteworthy that a strategy using a combination of both in vitro and in vivo technique would be preferable in future study of SLE. The application of genetic murine models of SLE will greatly advance this kind of work.
Conflict of interest: The authors declare that there is no conflict of interest regarding the publication of this article.
References
- 1. S, Zhao., Wang, Y., Liang, Y., et al. microRNA-126 regulates DNA methylation in CD4+T cells and contributes to systemic lupus erythmatosus by targeting methyltransferase 1. (2010) Arthritis Rheumatism 63(5): 1376-86
- 2. Dia, R., Zhang, Y., Khan, D., et al. Identification of a common lupus disease associated microRNA expression pattern in three different murine models of lupus. (2010) PLoS One 5(12): 14302.
- 3. Wen, P., Zhu, S., Yuan, M., et al. MicroRNA-21 and MicroRNA-148a+Lupus CD4 Contribute to DNA Hypomethylation in T Cells by Directly and Indirectly Targeting DNA Methyltransferase 1. (2010) J Immunol 184 (12): 6773-6781.
- 4. Zhang, J., Yang, B., Li, D., et al. Association of pre-microRNAs genetic variants with susceptibility with systemic lupus erythematosus. (2011) Mol Biol Rep 38(3): 1463–1468.
- 5. Chan, E.K., Satoh, M., Pauley, K.M. Contrast in Aberrant MicroRNA Expression in Systemic Lupus Erythematosus and Rheumatoid Arthritis: Is MicroRNA-146 All We Need? (2009) Arthritis Rheum 60(4): 912-915.
- 6. Bertsias, G., Cervera, R., Boumpas, T.D. Systemic Lupus Erythematosus: Pathogenesis and Clinical Features. 2012.
Pubmed || Crossref || Others
- 7. Lourenço, S.V., de Carvalho, F.R., Boggio, P. Lupus erythematosus: Clinical and histopathological study of oral manifestations and immunohistochemical profile of the inflammatory infiltrate. (2006) J Cut Pathol 34(7): 558-64.
- 8. Liu, C.C., Ahearn, J.M.The search for lupus biomarkers. (2009) Best Pract Res Clin Rheumatol 23(4): 507-523.
- 9. Luo, X., Tsai, L.M., Shen, N., et al. Evidence for microRNA-mediated regulation in rheumatic diseases. (2010) Ann Rheum Dis 69(1): 30-36.
- 10. Huang, Z., Huang, D., Ni, S., et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. (2010) Int J Cancer 127(1): 118-126.
- 11. Sanghamitra, B., Mitra, R., Maulik, U., et al. Research Development of the human cancer microRNA network. (2010) Silence 1(1): 6.
- 12. Mostert, B., Sieuwerts, A.M., Martens, J.W., et al. Diagnostic applications of cell-free and circulating tumor cell-associated miRNAs in cancer patients. (2011) Expert Rev Mol Diagn 11(3): 259-275.
- 13. Ceribelli, A., Satoh, M., Chan, E.K., et al. MicroRNAs in systemic rheumatic diseases. (2011) Arthritis Res Ther 13(4): 229.
- 14. Liang, D. N. S. MicroRNA involvement in lupus: the beginning of a new tale. (2012) Curr Opin Rheumatol 24(5): 489-498.
- 15. Asirvatham, A.J., Magner, W.J., Tomasi, T.B., et al. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. (2008) Mol Immunol 45(7): 1995-2006.
- 16. Stagakis, E., Iliopoulos, D., Boumpas, D.T., et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. (2011) Ann Rheum Dis 70(8): 1496-1506.
- 17. De-cai, Y.u., Qing-Guo, L.i., Yi-Tao, Ding., et al. Circulating MicroRNAs: Potential Biomarkers for Cancer. (2011) Int J Mol Sci 12(3): 2055–2063.
- 18. Li, Y., Li, W., Ouyang, Q., et al. Detection of lung cancer with blood microRNA-21 expression levels in Chinese population. (2011) Oncolo Lett 2(5): 991-994.
- 19. Garchow, B.G., Bartulos Encinas, O., Leung, Y.T., et al. silencing of microRNA-21 in vivo ameliorates autoimmune splenomegali in lupus mice. (2011) EMBO Mol Med (10): 605-15
- 20.Brase, J.C., Kuner, R., Sültman, H., et al. Serum microRNAs as non-invasive biomarkers for cancer. (2010) Mol Cancer 9: 306.
- 21. Gabor, G. I., Larissa, L., Edward, T., et al. Biomarkers in Systemic Lupus Erythematosus. (2004) Arthritis Rheumat.
- 22. Fenoglio, C., Cantoni, C., and De Riz, M. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. (2011) Neurosci Lett 504(1): 9-12
- 23. Ruan, Q., Wang, T. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic. (2011). Proc Natl Acad Sci 108(29): 12030-12035
- 24. Wu, F., Zikusoka, M. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. (2008) Gastroenterology 135(5): 1624-1635
- 25. Takagi, T., Naito, Y., Mizushima, K. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. (2010) J Gastroenterol Hepatol 1: S129-33.
- 26. Meisgen, F., Xu, N., and Wei, T. MiR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. (2012). Exp Dermatol 21(4): 312-314.
- 27. Mycko, M.P., Cichalewska, M., Machlanska, A. MicroRNA-301a regulation of a T-helper 17 immune response control autoimmune. (2012). Proc Natl Acad Sci 109(20): E1248-1257.
- 28. Zhu, H., Li, Y., Qu, S. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. (2012). J Clin Immunol 32(3): 514-522
- 29. Wang, H., Peng W, Ouyang, X. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. (2012) Transl Res 160(3): 198-206