Phage based diagnosis of bacterial infections
Vimlesh Gupta
Affiliation
Department of Veterinary Microbiology, College of Veterinary Science, GADVASU, Ludhiana, India
Corresponding Author
Hari Mohan Saxena, Department of Veterinary Microbiology, College of Veterinary Science, GADVASU, Ludhiana, India, Tel: +919417147813, E-mail: hmsaxena@yahoo.com
Citation
Saxena, H.M., et al. Covariation Phage based Diagnosis of Bacterial Infections. (2016) J Clin Trials Pathol Case Stud 1(1): 22- 25.
Copy rights
© 2016 Saxena, H.M. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Bacteriophage phage; Diagnosis; Bacterial infections; Bacterial diseases
Abstract
Accurate diagnosis and proper treatment of infectious diseases in the early stages are essential in preventing financial losses and patient suffering. Advanced diagnostic tests e.g. ELISA, CFT, PCR etc. are very costly, cumbersome, require sophisticated equipment and skilled personnel and cannot be employed at field level. Bacteriophages are viruses that specifically infect bacteria, replicate in it and lyse it. Lysis of the bacteria can be detected by using suitable indicator system. By employing bacteriophages along with indicator systems, accurate, easy and cost effective diagnosis of bacterial pathogens is possible in field conditions. Phage based diagnostic methods are cheap, simple and highly specific for bacterial diseases and are of bedside and penside applicability. Phage based diagnostics employing phage amplification and phage plaque method, reporter genes for luminescence, Fluorescence assays, MTT assay, Adenylate Kinase assay and ATP assay have been described here.
Introduction
Accurate diagnosis and proper treatment of infectious diseases in the early stages are essential in preventing financial losses and patient suffering. Various advanced diagnostic tests e.g. ELISA, CFT, PCR etc. are available but these are not employed in field because they are cumbersome, require very costly, sophisticated equipment and skilled personnel and can be performed in specialized labs only. Available ELISA and molecular assay based kits are very costly.
Bacteriophages
Bacteriophages are viruses that infect and replicate in prokaryotic cells. Phages kill between 4% - 50% of the bacteria produced every day, are driver of global geochemical cycles and a reservoir of the greatest genetic diversity on Earth[1]. Two independent observations made by[2,3] who described glassy transformations of micrococci colonies and antagonistic bacterial microbes, respectively, are seen as the onset of modern phage research.
In 1917 bacteriophages were officially discovered by d’Herelle, a French Canadian Microbiologist at the Pasteur Institute in Paris[4] who coined the name bacteriophages, derived from ‘bacteria’ and ‘phagein’ (Greek), which means ‘to eat’. He also introduced the term ‘plaque’ to describe the circular area of clearing caused by infection of a single phage on double-layered agar plates.
Phage based diagnostic assays
By employing bacteriophages along with suitable indicator systems, easy diagnosis of bacterial pathogens is possible in field conditions. The use of bacteriophage in assays for detecting bacteria was first reported over half a century ago when an assay to detect Salmonella using the phage Felix 01 was described by Cherry et al in 1954[5]. Bacteriophage based diagnosis is particularly useful when studying slow growing organisms such as Brucella spp., Mycobacterium tuberculosis and Mycobacterium avium subsp. Paratuberculosis[6]. The ability of bacteriophage to specifically infect, and lyse its host bacterium is to be exploited as a means of uniquely identifying target bacteria. After lysis by the specific lytic phage, release of specific contents of the bacteria can be detected by using suitable indicator system. Phage-based assays are particularly attractive since they are rapid, simple, and do not require the use of expensive equipment. Phage based diagnosis is easy because phages can be maintained at room temperature[7]. Observed that after lyophilization, phage retained its activity during storage for at least 20 months at 4°C. Phage is stable for 24 hours in broth at pH values from 6 to 8 at 37°C. The specificity of bacteriophage for their hosts has made them ideal tools for the classification, identification, and detection of bacteria.
Phage amplification and phage plaque method
Favrin et al[8] developed a novel assay that utilizes the normal infection cycle of bacteriophage SJ2 for detection of Salmonella enterica serovar Enteritidis in broth. It included 4 stages (i) capture and concentration of target cells by using Immuno Magnetic Separation (IMS) (ii) infection of the target bacterium with phage (iii) amplification and recovery of progeny phage and (iv) assay of progeny phage on the basis of their effect on a healthy population of host cells (signal-amplifying cells). The end point of the assay can be determined by using either fluorescence or optical density measurements. The detection limit of the assay in broth is less than 104 CFU/ml, and the assay can be performed in 4 to 5 h.
McNerney et al[9] reported that successful infection and replication of bacteriophages is indicative of the presence of viable bacteria. They described the development of a bacteriophage replication assay for the detection of Mycobacterium tuberculosis by using mycobacteriophage D29. Optimization of phage inoculation and incubation times allowed highly sensitive detection of M. bovis. Mythri and Samaga[10] collected three consecutive sputum samples. After decontamination and concentration techniques, samples were used for phage assay, cultured on Lowenstein-Jensen medium and smears were prepared. The overall sensitivity, specificity, PPV and NPV of phage assay when compared to LJ culture were 83.3%, 100%, 1.0 and 0.92, respectively. With respect to smear-negative specimens, the sensitivity was 50%.
Reporter phage based assays
Meighen[11] reported that bioluminescent bacteria, including Photobacterium, Vibrio, and Photorhabdus are capable of emitting light. The light-emitting reaction involves an intracellular luciferase. Bacterial luciferase is a heterodimeric enzyme and coded by the lux A and lux B genes. lux A and lux B genes have been used as reporter genes for bacterial detection. The principle of this detection method requires the introduction of the lux genes into the genome of a bacteriophage. The recombinant phages lack the intracellular biochemistry necessary for light production. However, infection of host bacteria by phage leads to the expression of phage genes and the additional lux gene within 30 to 50 minutes[12]. The result of phage infection is bioluminescent bacteria. The specificity of the detection method is determined by bacteriophage and host specificity and only viable cells are detected.
Hennes et al[13] used fluorescently stained bacteriophages to identify and enumerate specific strains of heterotrophic bacteria and cyanobacteria that were added to natural marine microbial communities. Chen and Griffiths[14] constructed recombinant bacteriophages specific for Salmonella spp and containing either lux AB or the entire lux operon. By employing a 6 hour preincubation step of this reporter phage and Salmonella, as few as 10 Salmonella cells per ml in the original sample could be detected. Loessner et al (1996) reported that by using the recombinant Listeria phage A511: luxAB, as few as 5 x 102 CFU/ml of tested Listeria cells could be detected after phage infection and two hours incubation at 20°C.
Goodridge et al[15] developed a fluorescent bacteriophage assay for the detection of E. coli O157:H7 in ground beef and raw milk samples. In this assay, fluorescently labelled phage AR1 specific for E. coli O157:H7 was used. Attachment of the labelled phage AR1 to the surface of E. coli O157:H7 could be visualized by epifluorescent microscopy or quantified by flow cytometry. The detection limits of this assay were approximately 1 × 103 CFU/ml in raw milk.
Edgar et al[16] reported biotinylation of engineered host-specific bacteriophage and conjugation of the phage to streptavidin-coated quantum dots. The method provides specific detection of as few as 10 bacterial cells per ml in experimental samples, with an approximately 100-fold amplification of the signal over background in 1 h.
MTT assay and other biochemical assays
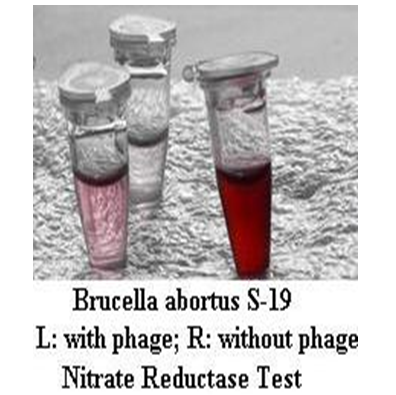
Gupta and Saxena[17] adapted MTT assay and nitrate reduction assay for use along with phage in detection of Brucella organisms. In phage based MTT assay, all the standard isolates of Brucella changed colour from yellow to purple upon addition of MTT after 6.5 hours of incubation of bacteria alone. However, in case of wells containing bacteria incubated with the specific brucellaphage, no colour change was observed after 6.5 hours of incubation because of the specific lysis of bacteria by the phage. In phage based Nitrate Reduction Test, red colour was noticed in the case of live Brucella isolates alone, but in case of Brucella incubated with phage for 8 hours, it remained colourless. The above mentioned tests can be suitably adapted to develop pen side kits for use with clinical samples after concentration of organisms in sample with positive serum and reduction of sample contamination using certain antibiotics.
Figure 1: Phage based Nitrate Reductase Test in small tubes.
Figure 2: Phage based MTT assay for Salmonella in small tubes.
Table 1: Confirmation of Salmonella by cost effective phage mediated lysis and costly PCR.
| Isolate of Salmonella | PCR positivity | Lysis by phage |
|---|---|---|
| 37s | + | + |
| 38s | + | + |
| 31s | + | + |
| 59s | + | + |
| 71s | + | + |
| 80s | + | + |
| 96s | + | + |
| 12s | + | + |
| 49s | + | + |
ATP and AK assays
The ATP bioluminescence reaction is catalysed by the enzyme luciferase, and requires a series of substrates including ATP (adenosine triphosphate) and luciferin. Here ATP functions to link the enzyme luciferase to its substrate luciferin.
Sanders (1995) used bacteriophage for specific lysis of Listeria monocytogenes, and the subsequent detection of an increase in the level of released bacterial ATP. This method enabled to rapidly and selectively identify 2.5 x 105 cells of L. Monocytogenes in the presence of one other Listeria species within 80 minutes after the addition of bacteriophage. Murphy et al[18] also investigated the phage-mediated adenylate kinase (AK) assay. This assay was claimed to be more sensitive than the phage-mediated ATP assay.
Blasco et al[19] used bacteriophages to cause specific lysis of bacteria and developed sensitive and rapid assay for the specific detection of bacteria using Escherichia coli and Salmonella Newport as the test organisms. The release of cell contents was measured by ATP bioluminescence. Increased sensitivity was obtained by focusing on the bacteria’s adenylate kinase (AK) as the cell marker instead of ATP as conventionally used. Fewer than 103 E. coli cells could be readily detected in less than 1 h. Salmonella Newport assays, although as sensitive, were slower and took up to 2 h.
Minikh et al[20] used wild type T4 bacteriophage and recombinant T4 bacteriophages displaying biotin binding peptide and cellulose binding module on their heads. These phages were immobilized on nano-Aluminum fiber-based filter, streptavidin magnetic beads and microcrystalline cellulose, respectively. Infectivity of the immobilized phages was investigated by monitoring the phage-mediated growth inhibition of bioluminescent E. coli B and cell lysis using bioluminescent ATP assay. Excess of interfering microflora at levels 60-fold greater than the target organism did not affect the results when bacteriophage was immobilized on the filter prior to concentration of bacterial cells.
Saxena and Gupta[21] exploited the phenomena of ATP release on phage mediated lysis of bacteria and ATP catalyzed luciferase-luciferin reaction to develop a novel assay for diagnosis of Brucellosis from clinical samples employing the ATP Determination kit (Invitrogen Detection Technologies). Mean luminescence was 1616.333 ± 662.608 for Brucella positive uterine secretions alone and 18507 ± 3327.018 for phage treated positive samples. There was a very significant difference (P < 0.01) between the two values. The average increase in luminescence was 10.03 folds. The mean luminescence of negative samples treated with phage was 700.333 ± 364.664. The difference between the luminescence of phage treated positive and negative samples was very significant (P < 0.01). The positive/negative status of animals from which the samples were derived was confirmed by Rose Bengal Plate Test and ELISA. The novel assay is simple, easy, accurate and field applicable assay for Brucellosis.
Thus, phage based methods offer cheap, simple and highly specific diagnostic tests for bacterial diseases which are of bedside and penside applicability.
References
- 1. Suttle, C.A. Viruses in the sea. (2005) Nature 437(7057): 356-361.
- 2. Twort, F.W. An investigation on the nature of ultramicroscopic viruses. (1915) Lancet 186(4814): 1241-1243
- 3. d'Hérelle, F. Sur un microbe invisible antagoniste des Bacilles dysentériques. (1917) Critical Reviews Academic Science Paris 165: 373-375.
- 4. d’Herelle, F. The bacteriophage. (1949) Science News 14: 44-59.
- 5. Cherry, W.B., Davis, B.R, Edwards, P.R. et al. A simple procedure for the identification of the genus Salmonella by means of a specific bacteriophage. (1954) J Lab Clin Med 44(1): 51-55.
- 6. Stanley, E.C., Mole, R.J., Smith, R.J., et al. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. (2007) Appl Environ Microbiol 73(6): 1851-1857.
- 7. McDuff, C.R., Jones, L.M., Wilson, J.B. Characteristics of brucellaphages. (1961) J Bacteriol 83(2): 324-329.
- 8. Favrin, S.J., Jassim, S.A., Griffiths, M.W. Development and optimization of a novel Immunomagnetic Separation Bacteriophage Assay for detection of Salmonella enterica Serovar Enteritidis in broth. (2001) Appl Environ Microbiol 67(1): 217-224.
- 9. McNerney, R., Kambashi, B.S., Kinkese, J., et al. Development of a bacteriophage phage replication assay for diagnosis of pulmonary tuberculosis. (2004) J Clin Microbiol 42(5): 2115-2120.
- 10. Mythri, K.M., Samaga, M.P. Rapid diagnosis of pulmonary tuberculosis by phage assay in HIV positive patients. (2013) Int J Pharm Bio Sci 4(2): 257-262.
- 11. Meighen, E.A. Genetics of bacterial bioluminescence. (1994) Ann Rev Genet 28: 117-139.
- 12. Stewart, G.S.A.B., Williams, P. Lux genes and the applications of bacterial bioluminescence. (1992) J Gen Microbiol 138(7): 1289-1300.
- 13. Hennes, K.P, Suttle, C.A., Chan, A.M. Fluorescently labelled virus probes show that natural virus populations can control the structure of marine microbial communities. (1995) Appl Environ Microbiol 61(10): 3623-3627.
- 14. Chen, J., Griffiths, M.W. Luminescent Salmonella strains as real time reporters of growth and recovery from sublethal injury in food. (1996) Int J Food Microbiol 31(1-3): 27-43.
- 15. Goodridge, L., Chen, J.,Griffiths, M.W. Development and characterization of a fluorescent-bacteriophage assay for detection of Escherichia coli O157: H7. (1999) Appl Environ Microbiol 65(4): 1392-1404.
- 16. Edgar, R., McKinstry, M., Hwang, J., et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. (2006) Proc Natl Acad Sci U S A 103(13): 4841-4845.
- 17. Gupta, V., Saxena, H.M. Application of bacteriophage in the diagnosis of brucellosis Veterinary Practitioner (2013) 14(2) (supple. 1).
- 18. Murphy, M.J, Squirrell, D.J., Sanders, M.F., et al. The use of adenylate kinase for the detection and identification of low numbers of microorganisrns. In: Campbell A K, Kricka L J, and 16. P.E. Stanley (ed.), Bioluminescence and Cherniluminescence. (1996) 320-322. John Wiley and Sons, Chichester.
- 19. Blasco, R., Murphy, M. J., Sanders, M. F., et al. Specific assays for bacteria using phage mediated release of adenylate kinase. (1998) J Appl Microbiol 84(4): 661-666.
- 20. Minikh, O., Tolba, M., Brovko. L.Y., et al. Bacteriophage-based biosorbents coupled with bioluminescent ATP assay for rapid concentration and detection of Escherichia coli. (2010) J Microbiol Methods 82(2): 177-183.
- 21. Saxena, H.M., Gupta, V. A new bacteriophage based luminescence assay for diagnosis of brucellosis. (2015) J Vet Sci Technol 6: 6.