Phenotypic Copper Resistance in Aerobic Intestinal Bacteria from Children with Different Levels of Copper-Exposure
Antonio Sanchez-Valenzuelaa#, Aida Ripolla#, Maria-Carmen Turrientesa,g, Eva Moralesb,g, Mercedes Rodriguez-Banosa, Mariona Bustamantec,h,i,g, Martine Vrijheidc,h,g, Maria Jose Lopez-Espinosad,f,g, Sabrina Llopd,f,g, Loreto Santa-Marinae,f,g, Ana Jimenez-Zabalae,f,g, Ana Moreno Bofarulla,g, Rafael Cantona, Jordi Sunyerc,h,j,g, Teresa M. Coquea,g, Fernando Baqueroa,g*
Affiliation
- aDepartment of Microbiology, Ramon y Cajal University Hospital, IRYCIS, Madrid, Spain
- bInstitute for Health Research, IMIB, Virgen de la Arrixaca University Hospital, Murcia, Spain
- cCentre for Research in Environmental Epidemiology (CREAL), Barcelona, Spain
- dEpidemiology and Environmental Health Joint Research Unit. FISABIO-Universitat Jaume I-Universitat de Valencia, Spain
- ePublic Health Division of Gipuzkoa, Basque Government, Spain. BIODONOSTIA Health Research Institute, Spain
- fSpanish Project for Environment and Childhood Research (INMA), Sabadell, Spain
- gSpanish Consortium for Research on Epidemiology and Public Health (CIBERESP), Carlos III Health Institute, Madrid, Spain
- hUniversitat Pompeu Fabra UPF, Barcelona, Spain
- iCenter for Genomic Regulation (CRG), the Barcelona Institute of Science and Technology, Barcelona, Spain; IMIM, Hospital del Mar Medical Research Institute, Barcelona, Spain
- #A.S.V and A.R contributed equally to this work
Corresponding Author
Fernando Baquero, Department of Microbiology, Ramón y Cajal University Hospital, IRYCIS and Spanish Consortium for Research on Epidemiology and Public Health (CIBERESP), Carlos III Health Institute, Carretera de Colmenar, Km 9.100. 28034 Madrid, Spain, Tel: +34-913368330; E-mail: baquero@bitmailer.net
Citation
Baquero, F., et al. Copper Resistance in Aerobic Intestinal Bacteria from Children with Different Levels of Copper-Exposure. (2017) J Environ Health Sci 3(1): 1-13.
Copy rights
© 2017 Baquero, F. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Copper-resistance; E. coli; K. pneumoniae; Copper-exposure; Healthy children
Abstract
Copper is an environmental element, essential for life, to which humans are exposed by inhalation, hand-to-mouth contamination, or ingestion of food and water. In this work, bacterial copper susceptibility (amount of copper able to inhibit bacterial growth) of aerobic bacteria from the intestinal microbiota of healthy children in Spain was explored. To establish the possible effect of children’s exposure to copper in the selection of copper-resistant organisms, the prevalence of copper-resistance among bacteria isolated in stool samples of 233 children belonging to the INMA cohort of the Spanish Project for Environment and Childhood Research was studied. Stool samples were seeded into Szybalsky-type agar plates containing specific culture media and a gradient of copper sulphate (< 0.68 - 2.05 mM). Culture media was suitable for growth of Gamma-Proteobacteria (mostly Enterobacteriaceae), Enterococcus and Staphylococcus. Colonies growing at intermediate (IR: < 0.68 - 1.36 mM) and/or higher copper concentrations (HR: 1.36 - 2.05 mM) were characterized by MaldiTOF assays. Sixty different species of copper-resistant organisms were detected. For Enterobacteriaceae, HR colonies were detected in 64.5% of seeded fecal samples, mostly corresponding to genus Escherichia (77% of positive samples); Enterobacter (15%), Citrobacter (13%), and Klebsiella (4.7%). However, Escherichia coli populations have a significantly lower proportion of high copper resistant colonies (49%) than the ensemble of Klebsiella-Enterobacter- Citrobacter colonies (77%). In HR-colonies of Firmicutes, Enterococcus genus was found in 97.3% of fecal samples, predominantly E. faecium (86%, of the positive Enterococcus samples), E. faecalis (37%), and E. hirae (6.9%). No significant correlation was found between counts of HR-colonies and the copper concentrations found in dry hair of the children studied (10 - 30 mcg/g). Copper-resistant populations in the children’s intestine might have evolved in the copper-rich external environment.
Introduction
Copper, an essential micronutrient for most plant and animal species, is involved in the function of several enzymes and other proteins, and needed in a wide range of metabolic processes; conversely, high concentrations can be detrimental for health[1]. However, the clinical consequences of both low and high copper-levels in the human body are rarely observed, but it should be noted that the possible effects on human microbiome diversity have scarcely been explored[2].
Human exposure to copper mainly occurs through the ingestion of food. The most significant sources of the dietary copper are yeast breads, white potatoes, tomatoes, cereals, beef, liver, seafood (particularly shellfish), dried beans and lentils[3]. Also copper reaches humans by drinking water and inhalation of polluted air (linked to occupational exposures)[4]. Natural ingredients in the normal Western diet provide approximately 2 to 4 mg copper per day; about 0.6 to 1.6 mg is absorbed, from 0.5 to 1.2 mg is excreted in the bile, 0.1 to 0.3 mg passes directly into the bowel, and 0.01 to 0.06 mg appears in the urine[5]. Much of the copper that is absorbed is later excreted in the bile so that more than 90 percent of ingested copper is found in feces[6]. Note that natural food exposure might be eventually be increased by GRAS (“Generally Recognized as Safe”), including cuprous iodide in table salt, as nutrients and/or dietary supplements, as antiseptic in paper and paperboard products used in food packaging, and growth promoters in foodborne animals[7].
Dietary supplements including copper in farm animals might modify the bacterial microbiota, frequently reducing Gram-negative facultative (coliform) organisms[8]. In fact poultry coliforms (Enterobacteriaceae, Proteobacteria) are more susceptible to copper than gram positives as Lactobacillus (Firmicutes)[9]. Early studies suggested a relative tolerance of lactobacilli and Escherichia coli to copper as dietary supplement[10].
The possible effects of copper on the normal microbiome were not listed among the biological effects of copper human exposure[4]. Most importantly, copper-resistance genes are frequently located in the same genetic platforms as genes encoding resistance to other metals (zinc, mercury, arsenic) and antibiotic resistance genes, and therefore the conditions favoring copper-resistance in the individual might influence antibiotic-resistance and vice versa[11,12].
The aim of this work is to investigate the frequency of copper-resistance in bacterial populations from the major facultative Proteobacteria and Firmicutes taxa in the intestine of healthy children with different documented levels of copper in dry hair. We studied the prevalence of bacterial copper-resistance in fecal samples of 233 children belonging to the INMA cohort. Samples were seeded in selective culture media containing gradients of intermediate (< 0.68 - 1.36 mM) or high (1.36 - 2.05 mM) copper concentrations. This range of concentrations, lower than are generally studied, permits exploration of the effects of copper when it starts influencing growth of bacterial populations. This semi-quantitative approach allowed a complete detection of the target intestinal population able to form colonies in a continuum of concentrations, thus revealing different levels of copper-resistance. With this technique we reached our objectives addressed to obtain: 1) the proportion of intestinal samples containing the different copper-resistant species; 2) the frequency of copper-resistance among isolates of different bacterial species, and 3) the correlation between copper exposure and the density or frequency of copper resistant bacteria.
There was an almost absolute absence of data concerning the levels of exposure of normal individuals to copper under the standard conditions of developed countries, and the effects that such exposure produce in the human intestinal microbiota. Our research was focused on ascertaining this correlation by measuring changes in the proportion of copper-resistant bacteria (selection for resistance) in children with different levels of copper resistance. At the same time, our work for the first time documents the frequency of copper-resistant bacteria in clinically-relevant bacteria of the intestinal microbiota of healthy children.
Materials and Methods
Study population and sample collection
The prevalence of copper-resistance in fecal samples was studied in 233 healthy children (1 - 8 years old), randomly selected from different Spanish towns and regions (Guipuzkoa, Sabadell and, Valencia), belonging to the cohort of Spanish Project for Environment and Childhood Research (INMA, http://www.proyectoinma.org/). The study was approved by the regional ethical committees of each regional cohort and all the parents of the children signed written informed consent before beginning the study. A total of 166 fecal samples (at least 10 g each, obtained by natural defecation) were obtained in Sabadell and Guipuzcoa, applying the MetaHit sampling protocol[13]. In children from Valencia, 67 rectal swabs (Conda Laboratories, Torrejon, Madrid, Spain) samples were obtained. In all the cases, samples were immediately frozen and transported to the recipient laboratory where they were stored at -40°C.
Chemical analysis of copper concentration in children’s hair
Chronic exposure to copper was evaluated in 75 children by the standard procedure in our group for measuring the metal concentration in the hair[14]. A minimum of 10 mg of hair from occipital scalp was collected for each child, placing samples in a plastic bag and storing them at room temperature until their dispatch. Copper concentrations in hair were obtained by using an atomic absorption spectrometer (AMA-254 analyzer from the Leco Corp. Praha, Czech Republic) at the Environmental Chemistry Department (IIQAB) of the CSIC (Barcelona, Spain).
Stool samples
Stool samples included feces (solid fecal material, fecal samples) and rectal swabs. For feces, one gram was homogenized by vortexing into 5 ml of sterile saline (9 g of sodium chloride salt per litre). One-tenth dilution was performed in saline, and 200 μL were seeded (rolling beads) into the copper-containing culture media. This dilution was plated on Petri square (12 x 12 cm, 144 cm²) gradient plates, containing culture agar media with a copper sulfate gradient (CuSO4*5H2O) ranging from 0 to 512 μg/ml (0 - 2.05 mM). McConkey Agar medium (Becton Dickinson BBLTMMcConkey Agar, Ref. 211387) was used for Enterobacteriaceae (Proteobacteria) recovery, M-Enterococcus Agar (Becton Dickinson DifcoTM m-Enterococcus Agar, Ref. 274620) for selected aerobic Firmicutes recovery, and Mannitol Salt Agar (Conda, Ref. 1062.00). Due to the expected differences in growth rates between these organisms, McConkey plates were incubated at 37°C for 24 h, and for M-Enterococcus and Mannitol Salt agar (MSA) plates for 48 h.
For rectal swabs, the procedure was as following: the swab was suspended in 3 ml of Brain Heart Infusion (Conda, Ref 1400.00), and incubated for 24 h at 37°C. From this culture, 1.5 ml was centrifuged at 6,000 rpm for 6 minutes, and the pellet was re-suspended in 1 ml saline, and the optical density adjusted at 0.5 McFarland. 200 μL of this suspension were seeded (rolling beads) into the copper-containing gradient plates (see above paragraph), and incubated at 37°C for 24 h for McConkey plates, and for 48 h for M-Enterococcus and Mannitol Salt agar (MSA) agar plates.
Categorization of bacterial copper susceptibility
Due to the absence of well-defined cut-offs for establishing susceptibility or resistance to heavy metals (including copper) in natural bacterial isolates, and the expected wide gradient distribution of copper concentrations in nature, the evaluation of the inhibitory effect of copper was done as follows: the surface of the square plate was divided in three sectors, corresponding to copper concentrations of < 170 μg/ml (< 0.68 mM), 170 - 340 μg/ml (0.68 - 1.36 mM) and 340 - 512 μg/ml (1.36 - 2.05 mM). All colonial morphotypes growing in the different sectors of the plates were considered for identification and differential counting. In cases of uncountable or very scarce growth along the gradient plate, the assay was repeated with a higher or less diluted inoculum to be able to detect individual colonies. This number of colonial morphotypes is referred to along the manuscript as “number of isolates”. Colony counts were performed in all three sectors of the plate. For each sample the total number of colonies in the sectors of the gradient plate was counted. The density (colony counts/ml of sample) of the bacteria belonging to the aerobic taxa Gamma-Proteobacteria and Firmicutes was plotted against the copper levels measured in the hair sample of the corresponding child.
Bacterial colony species identification was performed with a matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) method, using the Microflex LT mass spectrometer (Bruker Daltonics GmbH, Leipzig, Germany) with the FlexControl 3.0 and MALDI BioTyper 2.0 and 3.0 software programs.
Data analysis and statistics
For statistical analysis we used the software provided in the Social Science Statistics web site (www.socistatistics.com). When the p-value is not specifically presented, significant differences are considered if p < 0.01. The Pearson correlation (r) was obtained to analyze the plots of colony counts/ml in all three sectors of the gradient plates versus the copper levels measured in the hair sample of the corresponding child.
Results
Copper resistance in intestinal Gamma-Proteobacteria
A detailed account of all bacterial species of Gamma- Proteobacteria with copper-resistant bacteria is shown in (Table 1). In the following paragraphs, the analysis of the frequency of recovery of these bacteria in stool samples and fecal swabs, and the proportion of copper resistant organisms in the different taxa is presented.
Table 1: Identified species containing organisms with high resistance to copper, (MIC 340 - 512 μg/ml, or 1.36 - 2.05 mM) in the INMA cohort (Spain).
| Phylum | Genus | Species | Number |
|---|---|---|---|
| Proteobacteria | Escherichia (211) | E. coli (210) | 105 |
| E. hermanii (1) | 0 | ||
| Enterobacter (20) | E. asburiae (2) | 2 | |
| E. cloacae (18) | 15 | ||
| Citrobacter (19) | C. braakii (5) | 2 | |
| C. freundii (13) | 11 | ||
| C. youngae (1) | 1 | ||
| Klebsiella (14) | K. oxytoca (9) | 4 | |
| K. pneumoniae (4) | 3 | ||
| K. variicola (1) | 1 | ||
| Raoultella (6) | R. ornithinolytica (3) | 1 | |
| R. planticola (3) | 3 | ||
| Pseudomonas (5) | P. fragi (1) | 1 | |
| P. monteilli (1) | 1 | ||
| P. plecogiossicida (1) | 1 | ||
| P. putida (2) | 2 | ||
| Acinetobacter (2) | A. calcoaceticus (1) | 1 | |
| A. junii (1) | 1 | ||
| Salmonella (1) | Salmonella sp. (1) | 1 | |
| Kluyvera (1) | K. cryocrescens (1) | 1 | |
| Hafnia (1) | H. alvei (1) | 1 | |
| Comamonas (1) | C. kerstersii (1) | 1 | |
| Firmicutes | Enterococcus(397) | E. faecium(246) | 196 |
| E. faecalis(101) | 72 | ||
| E. hirae(15) | 11 | ||
| E. durans(14) | 8 | ||
| E. casseliflavus (7) | 2 | ||
| E. avium (6) | 3 | ||
| E. mundtii (3) | 1 | ||
| E. gallinarum (2) | 2 | ||
| E. gilvus (1) | 0 | ||
| E. raffinosus (1) | 0 | ||
| E. thailandicus (1) | 0 | ||
| Streptococcus (21) | S. constellatus (1) | 0 | |
| S. equinus (1) | 0 | ||
| S. pasteurianus (3) | 1 | ||
| S. salivarius (16) | 1 | ||
| Lactobacillus (11) | L. paracasei (6) | 1 | |
| L. plantarum (3) | 2 | ||
| L. sakei (2) | 0 | ||
| Pediococcus (8) | P. pentosaceus (8) | 6 | |
| Aerococcus (1) | A. viridans(1) | 1 | |
| Staphylococcus (67) | S. aureus (18) | 5 | |
| S. epidermidis (22) | 7 | ||
| S. hominis (10) | 3 | ||
| S. arlettae (3) | 1 | ||
| S. saprophyticus (3) | 3 | ||
| S. xylosus (3) | 3 | ||
| S. cohnii (2) | 1 | ||
| S. equorum (2) | 2 | ||
| S. haemolyticus (2) | 2 | ||
| S. pasteuri (1) | 1 | ||
| S. succinus(1) | 1 |
Proportion of intestinal stool samples containing the different copper-resistant species (Table 2)
Fecal samples: Colonies highly resistant to copper (MIC 340 - 512 μg/ml, or 1.36 - 2.05 mM) were detected in 107 cases from 166 stool samples (64.5%), corresponding to species of the genus Escherichia (in 77.6% of the samples, 83/107), Enterobacter (15.0%, 16/107), Citrobacter (13.1%, 14/107), Klebsiella (4.7%, 5/107), Pseudomonas (3.7%, 4/107), Raoultella (1.9%, 2/107), Acinetobacter (1.9%, 2/107), Salmonella (0.9%, 1/107), Kluyvera (0.9%, 1/107), Hafnia (0.9%, 1/107), Comamonas (0.9%, 1/107), and other non-identified organisms (3.7%, 4/107). In 58.9% (63/107) of the samples only one resistant species was detected, but 28.0% (30/107) had two resistant species, 9.3% (10/107) three resistant species and 1.9% (2/107) of the samples contained four highly resistant species. Colonies with growth in a copper concentration of 170 - 340 μg/ (0.68 - 1.36 mM, intermediate section of the Petri gradient plate) but not beyond (note that these samples might also contain different colonies from other species growing at higher concentrations) were found in most of the samples (90.4 %, 150/166). They were identified as species of the genera Escherichia (in 90.7% of the samples, 136/150), Citrobacter (12.0%, 18/150), Enterobacter (10.7%, 16/150), Klebsiella (5.3%, 8/150), Raoultella (2.7%, 4/150), Acinetobacter (1.3%, 2/150), Salmonella (0.7%, 1/150), Kluyvera (0.7%, 1/150), Hafnia (0.7%, 1/150), Comamonas (0.7%, 1/150), and non-identified organisms (4.7%, 7/150).
Table 2: Gram negative (Gamma-Proteobacteria) isolates obtained in McConkey agar media supplemented with copper at high (340-512 μg/ml) and intermediate concentrations (170-340 μg/ml). Proportions (per sample and per species) from feces and rectal swabs are detailed in the table. Note that the same sample might contain different resistant bacteria species (the total of positive samples per microorganism might exceed the total number of positive samples).
| Per sample (166 feces, 67 swabs) | Per bacterial isolates (288 from feces, 140 from swabs) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growing at high Cu++ | Growing at intermediate Cu++ | Growing at high Cu++ | Growing at intermediate Cu++ | ||||||||||||||
| Feces | Swabs | Feces | Swabs | Feces | Swabs | Feces | Swabs | ||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| Samples with R strains | 107 | 64.5 | 65 | 95.6 | 150 | 90.4 | 65 | 95.6 | Resistant isolates | 163 | 56.6 | 132 | 94.3 | 271 | 94.1 | 140 | 100.0 |
| Escherichia coli | 83 | 77.6 | 49 | 75.4 | 136 | 90.7 | 51 | 78.5 | Escherichia coli | 105 | 64.4 | 71 | 53.8 | 199 | 73.4 | 77 | 55.0 |
| Escherichia hermanii | 0 | 0.0 | 1 | 1.5 | 0 | 0.0 | 1 | 1.5 | Escherichia hermanii | 0 | 0.0 | 1 | 0.8 | 1 | 0.4 | 1 | 0.7 |
| Enterobacter cloacae | 14 | 13.1 | 17 | 26.2 | 14 | 9.3 | 18 | 27.7 | Enterobacter cloacae | 15 | 9.2 | 23 | 17.4 | 15 | 5.5 | 24 | 17.1 |
| Enterobacter aerogenes | 0 | 0.0 | 2 | 3.1 | 0 | 0.0 | 2 | 3.1 | Enterobacter aerogenes | 0 | 0.0 | 2 | 1.5 | 0 | 0.0 | 2 | 1.4 |
| Enterobacter asburiae | 2 | 1.9 | 1 | 1.5 | 2 | 1.3 | 1 | 1.5 | Enterobacter asburiae | 2 | 1.2 | 1 | 0.8 | 2 | 0.7 | 1 | 0.7 |
| Citrobacter freundii | 11 | 10.3 | 2 | 3.1 | 13 | 8.7 | 2 | 3.1 | Citrobacter freundii | 11 | 6.7 | 3 | 2.3 | 13 | 4.8 | 3 | 2.1 |
| Citrobacter braakii | 2 1 1.5 | 1.9 | 1 | 1.5 | 4 | 2.7 | 1 | 1.5 | Citrobacter braakii | 2 | 1.2 | 3 | 2.3 | 4 | 1.5 | 3 | 2.1 |
| Citrobacter youngae | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Citrobacter youngae | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Klebsiella pneumoniae | 2 | 1.9 | 13 | 20.0 | 2 | 1.3 | 14 | 21.5 | Klebsiella pneumoniae | 3 | 1.8 | 15 | 11.4 | 2 | 1.1 | 16 | 11.4 |
| Klebsiella oxytoca | 4 | 3.7 | 3 | 4.6 | 5 | 3.3 | 3 | 4.6 | Klebsiella oxytoca | 4 | 2.5 | 3 | 2.3 | 7 | 2.6 | 3 | 2.1 |
| Klebsiella variicola | 1 | 0.9 | 2 | 3.1 | 1 | 0.7 | 2 | 3.1 | Klebsiella variicola | 1 | 0.6 | 2 | 1.5 | 1 | 0.4 | 2 | 1.4 |
| Kluyvera cryocescens | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Kluyvera cryocescens | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Hafnia alvei | 1 | 0.9 | 1 | 1.5 | 1 | 0.7 | 1 | 1.5 | Hatniaspp | 1 | 0.6 | 1 | 0.8 | 1 | 0.4 | 1 | 0.7 |
| Proteusspp | 0 | 0.0 | 1 | 1.5 | 0 | 0.0 | 1 | 1.5 | Proteusspp | 0 | 0.0 | 1 | 0.8 | 0 | 0.0 | 1 | 0.7 |
| Morganellaspp | 0 | 0.0 | 1 | 1.5 | 0 | 0.0 | 1 | 1.5 | Morganellaspp | 0 | 0.0 | 1 | 0.8 | 0 | 0.0 | 1 | 0.7 |
| Raultellaspp | 2 | 1.9 | 0 | 0.0 | 4 | 2.7 | 0 | 0.0 | Raultella(4 species) | 4 | 2.5 | 0 | 0.0 | 6 | 2.2 | 0 | 0.0 |
| Comamonas kerstersii | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Comamonas kerstersii | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Salmonellaspp | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Salmonellaspp | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Acinetobacter junii | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Acinetobacter junii | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Acinetobacter calcoaceticus | 1 | 0.9 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | Acinetobacter calcoaceticus | 1 | 0.6 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 |
| Pseudomonasspp | 4 | 3.7 | 0 | 0.0 | 4 | 2.7 | 0 | 0.0 | Pseudomonasspp (4 species) | 5 | 3.1 | 0 | 0.0 | 5 | 1.8 | 0 | 0.0 |
| Non-identified | 4 | 3.7 | 4 | 6.2 | 7 | 4.7 | 4 | 6.2 | Non-identified | 4 | 2.5 | 4 | 3.0 | 7 | 2.6 | 4 | 2.9 |
Rectal swabs: Using enriched cultures from rectal swabs, the recovery of colonies highly resistant to copper was increased in relation to that obtained with fecal samples (65/68; 95.6% vs in 107/166; 64.5% ). Colonies corresponded to Escherichia (in 76.9% of the samples, 50/65), Enterobacter (30.8%, 20/65), Klebsiella (27.7%, 18/65), Citrobacter (4.6%, 3/65), Hafnia (1.5%, 1/65), Morganella (1.5 %, 1/65), and Proteus mirabilis (1.5%, 1/65), and non-identified (6.2%, 4/65). Intermediate copper-resistant colonies were found in 65 out of 68 samples (95.6%), containing Escherichia (80%, 52/65, Enterobacter (32.3%, 21/65), Klebsiella (29.2%, 19/65), Citrobacter (4.6%, 3/65), Hafnia, Morganella and Proteus (1.5 %, 1/65 each one), and non-identified (6.2%, 4/65). Species of Raoultella, Salmonella, Acinetobacter, Pseudomonas or Comamonas were not recovered from rectal swabs.
Frequency of copper-resistance among isolates of different bacterial species and genera (Table 2)
Fecal samples: The frequency of strains (all colonial morphotypes in all stool fecal samples considered) with high copper-resistance varied among the different bacterial genera; in Escherichia (105 Cu-R among 211 Escherichia colonies, 105/211, 49.8%), in Enterobacter (17/20, 85%); in Citrobacter (14/19, 73.68%), in Klebsiella (8/14, 57.14%), Raoultella (4/6, 66.7%), Pseudomonas (5/5), Acinetobacter (2/2), Salmonella (1/1), Kluyvera (1/1), Hafnia (1/1), Comamonas (1/1), and non-identified 4/9). In strains with high copper-resistance, Escherichia strains were less frequently resistant than Citrobacter (p < 0.05), Enterobacter (p < 0.05), and Klebsiella (p = 0.05). Considering all 70 isolates of non-E. coli predominantly environmental species, high copper resistance reaches 77.14%, and the proportion of copper-resistance among all 211 E. coli (predominantly enteric) was significantly lower, 48.81 % (p < 0.001). The frequency of strains able to grow at intermediate copper concentrations (in a proportion, also in higher ones, see preceding paragraph) were: Escherichia (200/211, 94.8%), Enterobacter (18/20, 90%), Klebsiella (11/14, 78.57%) and Citrobacter, Acinetobacter, Comamonas, Kluyvera, Pseudomonas, Raoultella, Salmonella and non-identified (100%).
Rectal swabs: In the case of rectal swabs, the frequency of strains with high copper-resistance was: Escherichia (72/77, 93.50%), Klebsiella (18/19), 94.74%, and Enterobacter (13/13, 100%), Citrobacter, Hafnia, Morganella and Proteus (100%). In summary, in the case of E. coli (211 strains) a mean of 94.8% of the strains (colony morphotypes) were able to grow in the range 170 - 340 μg/ml, and a mean of 49.8% were growing at higher copper concentrations (340 - 512 μg/ml). For Enterobacter, Citrobacter, and Klebsiella, means of 90, 100, 78.57%, respectively survived at the range 170 - 340 μg/ml, and means of 85, 73.68 and 57.14% respectively survived at 340 - 512 μg/ml.
Copper resistance in intestinal Firmicutes
A detailed account of all bacterial species of Firmicutes with copper-resistant bacteria is shown in Table 1. In the following paragraphs, the analysis of the frequency of recovery of these bacteria in stool samples and fecal swabs, and the proportion of copper resistant organisms in the different taxa is presented.
Proportion of intestinal samples containing the different copper-resistant species (Tables 3a, 3b)
Fecal samples: In M-Enterococcus Agar, at high copper concentrations (340 - 512 μg/ml, or 1.36 - 2.05 mM), resistant colonies were identified in 149 out of 166 fecal samples (89.8%). Enterococcus genus was found in 145 out of 149 samples (97.3%). Within Enterococcus, the predominant species were E. faecium (86.2%, 125/145 positive Enterococcus samples), E.faecalis (37.24%, 54/145), E. hirae (6.9%, 10/145) and E. durans (5.5%, 8/145). Other resistant Enterococcus species were: E. avium (6.9%), E. casseliflavus (1.4%), E. gallinarum (1.4%) and E. mundtii (0.7% of Enterococcus). Other resistant species were Pediococcus pentosaceus (4% of samples), Lactobacillus sp (2%), Streptococcus sp (1.3%), Aerococcus viridans (0.7%). In M-Enterococcus plates, 27.5% (41/149) of these samples showed only one resistant species, 34.9% (52/149) had two resistant species, 19.5% (29/149) three resistant species, 14.8% (22/149) four resistant species and 0.7% (1/149) five resistant species of Firmicutes. In MSA Agar plates, 40 out of 166 stool samples (24.1%) presented highly resistant isolates, and 27 out of 40 (67.5%) corresponded to Staphylococcus genus. Within Staphylococcus, eleven different species were identified: S. epidermidis (25.9%, 7/27 positive Staphylococcus samples), S. aureus (18.5%), S. hominis (11.1%), S. saprophyticus (11.1%), S. haemolyticus (7.4%), S. equorum (7.4%), S. xylosus (7.4%), S. succinus (3.7%), S. pasteuri (3.7%), S. cohnii (3.7%), and S.arlettae (3.7% of the total of Staphylococcus). Lactobacillus sp was identified in 7 samples (17.5%, 7/40), Aerococcus viridans in 1 sample (2.5%, 1/40) and non-identified microorganisms in 9 samples (22.5%, 9/40). In MSA plates, 70% (28/40) of the samples with resistant isolates contained 1 resistant isolates.
In M-Enterococcus Agar at intermediate copper concentrations colonies were recovered in 163 of 166 stool samples studied (98.2%). Enterococcus genera were found in 158 of 163 samples (96.9%). Within the genus Enterococcus, the species found were E. faecium (85.4%, 135/158), E. faecalis (45.6%, 72/158), E. hirae (6.96%, 11/158) and E. durans (6.96%, 11/158). E. casseliflavus (n = 5), E. avium (2.1%), E.mundtii (1.9%), and E. gallinarum (1.3%). Lactobacillus sp was found in 7/163 total samples (4.3%), Pediococcus pentosaceus (4.3%), Streptococcus sp (3.1%), and Aerococcus viridans (0.6%). In MSA Agar plates, 73 of 166 stool samples (44.0%) presented bacterial growth at high copper concentrations, and 46 of these 73 (58.9%) were positive for Staphylococcus. The species identified inside the genus Staphylococcus were: S. epidermidis (39.1%, 18/46), S. aureus (19.6%, 9/46), S. hominis (17.4%), S saprophyticus (6.5%), S. haemolyticus (4.3%), S. xylosus(4.3%), S. equorum (4.3%), S. succinus (2.2%), and S. pasteuri (2.2%). Two species of Lactobacillus were identified in 7 samples (9.6%, 7/73), and Aerococcus in 1 sample (1.4%, 1/73). Two resistant isolates were detected in 25% (10/40) of samples.
Rectal swabs: In the case of enriched rectal swabs, at high copper concentrations, in M-Enterococcus Agar, 58 of the 67 samples (86.57%) contained high copper-resistant strains and 91.38% (53/58) had Enterococcus strains. Inside the genus Enterococcus, E. faecium accounted for 69.81% (37/53) E. faecalis for 39.6% (21/53), E. hirae for 9.4% (5/53), and E. durans for 1.9% (1/53). Only 13 samples (19.1%) were positive in MSA Agar in rectal swabs for highly resistant bacteria, corresponding in a 76.9% (10/13) to Staphylococcus species; S. aureus (n = 5), S. epidermidis (n = 3), S. warneri (n = 1) and S. hominis (n = 1). Non identified species were found in 30.8% (4/13) of the rectal swabs. A lower proportion of high copper-resistant isolates per sample were identified in the rectal swabs if compared with stool samples in both M-Enterococcus and MSA culture media, the percentages being of one Cu-resistant isolate per sample 46.55 (27/58) and 61.54% (8/13); two resistant isolates per sample in 43.1 (25/58) and 30.76% (4/13) and three resistant isolates were identified in 8.62 (5/58) and 0.0%, respectively).
Swab enriched samples inoculated in M-Enterococcus agar plates provided growth at intermediate copper concentrations in 63 out of 68 samples (94%); in 48 of them (76.2%) growth corresponded to Enterococcus strains. Inside Enterococcus, 77.1% (37/48) corresponded to E. faecium, 50.0% (24/48) for E. faecalis, 10.4 % to E. hirae, and 2.1 % to each one of the species E. durans, E. avium, E. casseliflavus, and E. mundtii. Copper-resistant Lactobacillus and Pediococcus were both recovered in 4.3% of the total of samples. Swab enriched samples inoculated in MSA Agar provided growth at intermediate copper concentrations in 21/67 (30.9%); Staphylococcus species were found in 13 of them (61.9%). Among Staphylococcus, S. epidermidis accounted for 53.8 %, S. aureus for 31.8%, and S. hominis and S. warnerii for 7.7%. Lactobacillus sp was found in 14.3% of the swab samples (3/21).
Table 3a: Gram positive (Firmicutes) aerobic isolates obtained in M-Enterococcus medium supplemented with copper at high (340 - 512 μg/ml) and intermediate concentrations (170 - 340 μg/ml). Proportions (per sample and per species) from feces and rectal swabs are detailed in the table. In italics, numbers and percentages of species inside the genus Enterococcus. Note that the same sample might contain different resistant bacteria species (the total of positive samples per microorganism might exceed the total number of positive samples).
| Per sample (166 feces, 67 swabs) | Per bacterial isolates (460 from feces, 117 from swabs) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growing at high Cu++ | Growing at intermediate Cu++ | Growing at high Cu++ | Growing at intermediate Cu++ | ||||||||||||||
| Feces | Swabs | Feces | Swabs | Feces | Swabs | Feces | Swabs | ||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| Total samples | |||||||||||||||||
| Samples with R strains | 149 | 89.8 | 58 | 86.57 | 163 | 98.2 | 63 | 94.0 | Resistant isolates (R) | 325 | 70.7 | 93 | 79.5 | 410 | 89.1 | 106 | 90.6 |
| With R-Enterococcus | 145 | 97.3 | 53 | 91.38 | 158 | 96.9 | 48 | 76.2 | R-Enterococcus | 295 | 90.8 | 81 | 87.1 | 365 | 89.0 | 89 | 84.0 |
| E. faecium | 125 | 86.2 | 37 | 69.81 | 135 | 85.4 | 37 | 77.1 | E. faecium | 196 | 66.4 | 50 | 61.7 | 231 | 63.3 | 52 | 58.4 |
| E. faecalis | 54 | 37.2 | 21 | 39.62 | 72 | 45.6 | 24 | 50.0 | E. faecalis | 72 | 24.4 | 25 | 30.9 | 94 | 25.8 | 28 | 31.5 |
| E. durans | 8 | 5.5 | 1 | 1.89 | 11 | 7.0 | 11 | 2.1 | E. durans | 8 | 2.7 | 1 | 1.2 | 11 | 3.0 | 1 | 1.1 |
| E. hirae | 10 | 6.9 | 5 | 9.43 | 11 | 7.0 | 5 | 10.4 | E. hirae | 11 | 3.7 | 5 | 6.2 | 12 | 3.3 | 5 | 5.6 |
| E. avium | 3 | 2.1 | 0 | 0.00 | 4 | 2.5 | 1 | 2.1 | E. avium | 3 | 1.0 | 0 | 0.0 | 5 | 1.4 | 1 | 1.1 |
| E. casseliflavus | 2 | 1.4 | 0 | 0.00 | 5 | 3.2 | 1 | 2.1 | E. casseliflavus | 2 | 0.7 | 0 | 0.0 | 5 | 1.4 | 1 | 1.1 |
| E. gallinarum | 2 | 1.4 | 0 | 0.00 | 2 | 1.3 | 0 | 0.0 | E. gallinarum | 2 | 0.7 | 0 | 0.0 | 2 | 0.5 | 0 | 0.0 |
| E. mundtii | 1 | 0.7 | 0 | 0.00 | 3 | 1.9 | 1 | 2.1 | E. mundtii | 1 | 0.3 | 0 | 0.0 | 3.0 | 0.8 | 1 | 1.1 |
| R-Lactobacillus sp | 3 | 2.0 | 0 | 0.00 | 7 | 4.3 | 0 | 0.0 | R-Lactobacillus sp | 3 | 0.9 | 0 | 0.0 | 8 | 2.0 | 0 | 0.0 |
| R-Aerococcus viridans | 1 | 0.7 | 0 | 0.0 | 1 | 0.6 | 0 | 0.0 | R-Aerococcus viridans | 1 | 0.3 | 0 | 0.0 | 1 | 0.2 | 0 | 0.0 |
| R-Pedioc. pentosaceous | 6 | 4.0 | 0 | 0.00 | 7 | 4.3 | 0 | 0.0 | R-Pedioc. pentosaceous | 6 | 1.8 | 0 | 0.0 | 8 | 2.0 | 0 | 0.0 |
| R-Streptococcus sp | 2 | 1.3 | 0 | 0.0 | 5 | 3.1 | 0 | 0.0 | R-Streptococcus sp | 2 | 0.6 | 0 | 0.0 | 7 | 1.7 | 0 | 0.0 |
| R-Non-identified/contam? | 14 | 9.4 | 12 | 20.69 | 16 | 9.8 | 15 | 23.8 | R-Non-identified/contam? | 18 | 5.5 | 12 | 12.9 | 21 | 5.1 | 17 | 14.5 |
Table 3b: High and intermediate resistant Firmicutes obtained in MSA agar media supplemented with copper at high (340 - 512 μg/ml) and intermediate concentrations (170 - 340 μg/ml). Counts (per sample and per species) from feces and rectal swabs are detailed in the table.
| Per sample (166 feces, 67 swabs) | Per bacterial isolates (200 from feces, 48 from swabs) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High Cu++ resistant | Int. Cu++ resistant | High Cu++ resistant | Int. Cu++ resistant | ||||||||||||||
| Feces | Swabs | Feces | Swabs | Feces | Swabs | Feces | Swabs | ||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| Samples with resistance | 40 | 24.1 | 13 | 19.1 | 73 | 44.0 | 21 | 30.9 | Resistant isolates (R) | 48 | 24.0 | 15 | 31.3 | 149 | 74.5 | 24 | 50.0 |
| With R-Staphylococcus | 27 | 67.5 | 10 | 76.9 | 46 | 63.0 | 13 | 61.9 | R-Staphylococcus | 29 | 60.4 | 11 | 73.3 | 54 | 36.2 | 17 | 70.8 |
| S. epidermidis | 7 | 25.9 | 3 | 30.0 | 18 | 39.1 | 7 | 53.8 | S. epidermidis | 7 | 24.1 | 4 | 36.4 | 18 | 33.3 | 8 | 47.1 |
| S. aureus | 5 | 18.5 | 5 | 50.0 | 9 | 19.6 | 4 | 31.8 | S. aureus | 5 | 17.2 | 5 | 45.5 | 10 | 18.5 | 7 | 41.2 |
| S. saprolyticus | 3 | 11.1 | 0 | 0.0 | 3 | 6.5 | 0 | 0.0 | S. saprolyticus | 3 | 10.3 | 0 | 0.0 | 3 | 5.6 | 0 | 0.0 |
| S. haemolyticus | 2 | 7.4 | 0 | 0.0 | 2 | 4.3 | 0 | 0.0 | S. haemolyticus | 2 | 6.9 | 0 | 0.0 | 2 | 3.7 | 0 | 0.0 |
| S. hominis | 3 | 11.1 | 1 | 10.0 | 8 | 17.4 | 1 | 7.7 | S. hominis | 3 | 10.3 | 1 | 9.1 | 9 | 16.7 | 1 | 5.9 |
| S. xylosus | 2 | 7.4 | 0 | 0.0 | 2 | 4.3 | 0 | 0.0 | S. xylosus | 3 | 10.3 | 0 | 0.0 | 3 | 5.6 | 0 | 0.0 |
| S. equorum | 2 | 7.4 | 0 | 0.0 | 2 | 4.3 | 0 | 0.0 | S. equorum | 2 | 6.9 | 0 | 0.0 | 2 | 3.7 | 0 | 0.0 |
| S. succinus | 1 | 3.7 | 0 | 0.0 | 1 | 2.2 | 0 | 0.0 | S. succinus | 1 | 3.4 | 0 | 0.0 | 1 | 1.9 | 0 | 0.0 |
| S. pasteuri | 1 | 3.7 | 0 | 0.0 | 1 | 2.2 | 0 | 0.0 | S. pasteuri | 1 | 3.4 | 0 | 0.0 | 1 | 1.9 | 0 | 0.0 |
| S. warnerii | 0 | 0.0 | 1 | 10.0 | 0 | 0.0 | 1 | 7.7 | S. warnerii | 0 | 0.0 | 1 | 9.1 | 0 | 0.0 | 1 | 5.9 |
| S. cohnii | 1 | 3.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | S. cohnii | 1 | 3.4 | 0 | 0.0 | 2 | 3.7 | 0 | 0.0 |
| S. arlettae | 1 | 3.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | S. arlettae | 1 | 3.4 | 0 | 0.0 | 3 | 5.6 | 0 | 0.0 |
| R-Lactobacillus sp | 7 | 17.5 | 0 | 0.0 | 7 | 9.6 | 3 | 14.3 | R-Bacillus sp | 7 | 14.6 | 0 | 0.0 | 4 | 32.9 | 3 | 12.5 |
| R-Aerococcus viridans | 1 | 2.5 | 0 | 0.0 | 1 | 1.4 | 0 | 0.0 | R-Aerococcus viridans | 1 | 2.1 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 |
| R-Micrococcus luteus | 0 | 0.0 | 0 | 0.0 | 1 | 1.4 | 0 | 0.0 | R-Micrococcus luteus | 0 | 0.0 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 |
| R-Non-identified/suspected contaminants | 9 | 22.5 | 4 | 30.8 | 36 | 49.3 | 4 | 19.0 | R-Non-identified/suspected contaminants | 11 | 22.9 | 4 | 26.7 | 44 | 29.5 | 4 | 16.7 |
Frequency of copper-resistance among isolates of different bacterial species and genera (Tables 3a and 3b)
Fecal samples: As expected, most high copper-resistant colonies (325) from stool fecal samples recovered in M-Enterococcus plates (MIC 340 - 512 μg/ml, or 1.36 - 2.05 mM) corresponded to the genus Enterococcus (90.77%, 295/325). Inside Enterococcus, the proportion of E. faecium was 66.44% (196/295), and 24.40% (72/295 = for E. faecalis, and 6 other Enterococcal species (E. hirae, E. avium, E. casselliflavus, E. durans, E. gallinarum, and E. mundtii) accounted for the rest (9.2%). Other colonies belonged to the genera Pediococcus (1.85%, 6/325), Lactobacillus (0.92%, 3/325), Streptococcus (0.62%, 2/325), and Aerococcus viridans (0.31%, 1/325). In MSA media, 48 highly resistant colonies were recovered, 60.4% belonging to the genus Staphylococcus (29/48) and were represented by 9 different species; S. epidermidis n = 7, S. aureus n = 5, S. saprolyticus (n = 3), S. haemolyticus (n = 2), S. hominis (n = 3), S. xylosus (n = 3), S. equorum (n = 2), S. succinus (n = 1), S. pasteuri (n = 1), and S. cohnii (n = 1) and S. arletae ( n = 1). Bacillus sp was identified in 14.6% (7/48) and non-identified species in 29.2% of the isolates (14/48). Details of the species are shown in Table 3.
Intermediate-resistant isolates from M-Enterococcus agar seeded with stool samples were identified in 89.1% of the colonial morphotypes tested; most of them (89.1%) corresponded to Enterococcus. Within the genus, E. faecium was predominant (63.3% of the Enterococcus), followed by E. faecalis (25.8%), E. hirae (3.3%), and E. durans (3%). In MSA agar, intermediate-resistant isolates from stool samples accounted for 74.5% of the colonial morphotypes, the majority of them (36.2%) being Staphylococcus, mostly S. epidermidis (33.3% of them), followed by S. aureus (18.5%) and S. hominis (16.7%). We recovered an unexpected high number of isolates that corresponded to the genus Bacillus (B. subtilis, 20; B. flexus, 9; B. licheniformis, 5; B. megaterium and Bacillus sp, 4 each; B. mojavensis and B. pumilus, 3 for each y B. vallismortis[1].
Rectal swabs: In the rectal swabs, high copper-resistant isolates grown on the M-Enterococcus plates (MIC 340 - 512 μg/ ml, or 1.36 - 2.05 mM) corresponded to the genera Enterococcus (87.1%). As to the proportion of the resistant Enterococcus species, 50 isolates out of 81 (61.73%) were identified as E. faecium, 25 out of 81 (30.86%) were E. faecalis and 6 out of 81 (7.41%) corresponded with two other species (E. durans and E. hirae). In MSA plates, isolates from 4 different species of high-copper resistance were detected within the genus Staphylococcus (73.3%, 11/15). Details of the species are as follows: S. aureus (n = 5), S. epidermidis (n = 4), S. hominis (n = 1) and S. warnerii (n = 1) and 4 of 15 isolates (total of resistant strains) belonged to non-identified bacteria (26.7 %).
At intermediate copper concentrations 84% of the resistant isolates obtained in swabs inoculated on M-Enterococcus agar plates were Enterococcus (89/106) and 4 species were identified, E. faecium was the microorganism with the highest representation (58.4%, 52/89), followed by E. faecalis (31.5%, 28/89), and E. hirae (5.6% (5/89) of the resistant isolates. Finally, resistant isolates obtained at intermediate concentrations in enriched swabs seeded on MSA agar were mostly Staphylococcus (70.8%); within the genus, S. epidermidis predominated (47.1%), followed by S. aureus (41.2%), S. hominis and S. warnerii (4.9 % each). Species from Bacillus were found in 12.5% of the isolates.
Tables 2 and 3 provide a detailed description of the proportions (per sample and bacterial isolates) of aerobic Firmicutes obtained in M-Enterococcus Agar medium and Manitol- Salt-Agar medium supplemented with copper at high (340 - 512 μg/ml) and intermediate concentrations (170 - 340 μg/ml) of broth-enriched fecal swab samples.
Recovery of copper-resistant bacteria: feces versus rectal swabs
For epidemiological purposes, rectal swabs are much easier to obtain and to handle than feces. Only indirect comparisons of the recovery of copper-resistant bacteria in both types of samples can be made in our study, as each child only provided either feces or swabs. In the case of amma-Proteobacteria, culturing broth enriched swabs resulted in a significantly (p < 0.001) higher yield of positive samples for high-copper resistant bacteria (65/67, 97%) than direct feces plating (107/166, 64.45%). This difference disappears in recovery for intermediate-resistant bacteria.
The advantage of swabs does not apply for the detection of high- or intermediate copper-R E. coli, or Citrobacter, and stools are just as effective (not statistically different) as swabs. However, high- or intermediate copper-resistant Enterobacter or Klebsiella are more efficiently recovered (statistically significant difference, p < 0.001) in the swabs. Both fecal samples and rectal swabs yielded a comparable proportion (non-statistically significant) of positive samples for high- or intermediate copper-resistance. The taxon diversity in high- or intermediatecopper resistant species in plates seeded with fecal samples was higher (18 taxa) than that recovered after plating enriched swabs (14 taxa).
Unlike Gamma-Proteobacteria, samples inoculated in M-Enterococcus presented no statistical difference in the yield of positive samples for high-level or intermediate-level of copper resistance for the aerobic Firmicutes organisms recovered. As in the case of Proteobacteria, the diversity of Firmicutes with high- or intermediate- copper resistant taxa was higher on plates seeded with feces (13 taxa) than with enriched swabs (5 and 8 taxa, respectively).
Similarly, no statistical difference was detected in the recovery rate of high- or intermediate- copper-resistant strains in fecal or enriched swab samples in MSA agar. As in all former cases, the recovered taxon diversity was higher in the high- or intermediate- copper resistant species in plates seeded with fecal samples (14 and 12 taxa) than that recovered after plating enriched swabs (5 and 7 taxa respectively).
Copper resistance reflects bacterial lifestyle?
Globally we were able to identify 60 different species containing copper-resistant organisms, 22 species of Gamma Proteobacteria and 38 of aerobic Firmicutes (Table 1, and text for Bacillus isolates). The genus Escherichia, mostly represented by E. coli, is constituted by bacterial organisms well adapted to the intestine of the mammals, and is rare in the environment (except in a transient way in sewage, due to fecal contamination). On the contrary, all other Gamma-Proteobacteria detected in this work are able to sustain an environmental lifestyle, that is, are able to reproduce regularly outside animal hosts, frequently with lower optimal growth temperatures than E. coli. To a certain extent, some members of the “environmental” genus, as Klebsiella (and more rarely Enterobacter and Citrobacter) are eventually found in association with the vertebrate mucosas, but much less frequently than E. coli. We can expect a higher copper exposure in the free environments than in the humans (see Discussion), so that the more adapted intestinal organisms might be “protected” from environmental copper stress. Consistently with these differences in ecology and lifestyle, and using the list of well characterized highly-resistant isolates presented in Table 4, E. coli has a significantly lower proportion (p < 0.001) of high copper resistant strains (105/211, 49.76%) than the ensemble of environmental Gamma-Proteobacteria (54/70, 77.14%). This difference remains significant comparing E. coli with the ensemble of Klebsiella, Enterobacter, and Citrobacter (P value of 0.002).
In the case of Firmicutes, ecological and lifestyle differences are less obvious. In the case of Enterococcus, E. faecalis is more frequent than E. faecium in the human intestine, but both species are able to survive efficiently in the environment. Probably both species are exposed to environmental copper, and E. faecium is slightly more copper-resistant (79.27%) than E. faecalis (71.28%), but without statistical significance (p = 0.09). A similar difference was found comparing E. faecalis with all other Enterococci. In the case of Staphylococcus, the genus has very poor environmental representation; for instance, copper-resistance is not significantly different between the predominant species found, S. aureus and S. epidermidis.
Proportions and population densities of copper-resistant bacteria in the intestine
A quantitative analysis of the number of colonies obtained in each of the three sectors of the square plates containing low (L), intermediate (M), or high (H) copper concentrations (sector L, < 170 μg/ml or < 0.68 mM, 170-sector M, 170 - 340 μg/ml or 0.68 - 1.36 mM, sector H (340 - 512 μg/ml or 1.36 - 2.05 mM) was performed. Percentages of colonies in sector M and H with respect to those in sector L were estimated to offer an estimation of the effect of copper concentration intervals in the survival of bacterial populations, and hence of its possible selective effect. In the case of Gamma-Proteobacteria (Enterobacteriacae), the mean percentages of growth at M and H were, for E. coli, 52% and 11%; for Klebsiella, 59% and 31%, for Enterobacter, 69% and 26%, and for Citrobacter, 75% and 32%, respectively. These results show that “environmental Proteobacteria” are not only more frequently copper-resistant than E. coli, but also that proportion of the population able to tolerate medium and high copper concentrations is also higher (p < 0.01). For Firmicutes, in the case of E. faecium, the survivor’s frequency in the population in the sectors M and H, compared with the colony count in sector L, was 72% and 39%. For E. faecalis, the proportions were 71% and 22% and for E. hirae, 57% and 29%, respectively.
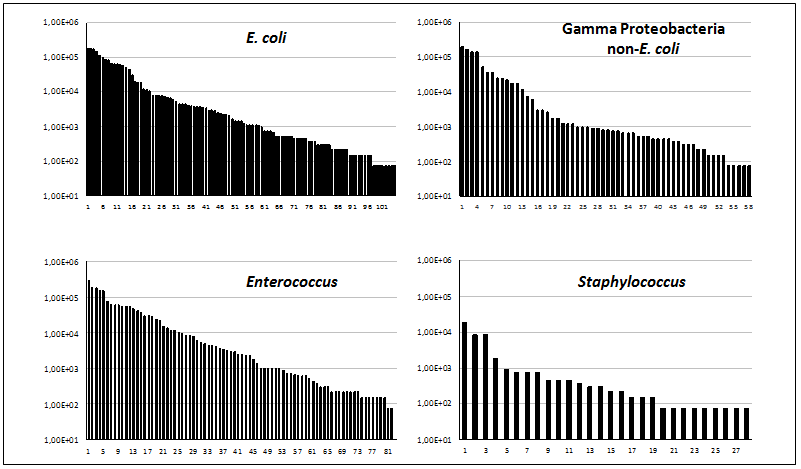
The total population size (bacterial counts/ml) of high copper-resistant colonies in fecal samples was reliably estimated for E. coli (103 samples), non-E. coli Proteobacteria (57 samples), Enterococcus (82 samples) and Staphylococcus (28 samples) (Figure 1). The proportion of samples containing ≥ 103 cells/ml of sample was higher in E. coli (58.25%) than in other Gamma-Proteobacteria (42.10%), which could correspond with the better intestinal adaptation of E. coli. This means that, even if E. coli is less frequently resistant to copper, the population density of copper resistant E. coli in the intestine surpasses other Gamma-Proteobacteria. In the aerobic Firmicutes, copper-resistant Enterococcus counted ≥ 10³ cells/ml in 62.19% of the samples, and Staphylococcus in only 14.28%. Considering that Enterococcus has high rates of copper-resistance, these results indicate that Enterococcus is the main quantitative contributor to high copper resistance in the children’s intestine. It should be noted that we were unable to recognize in the distributions presented in Figure 1 any sign of bimodal distribution, meaning that the abundance of copper-resistant bacteria is probably not the result of specific selective events. Lack of bimodality was also obtained in previous studies even using higher copper concentrations (4 - 16 mM)[14].
Figure 1: Viable counts (cfu/ml) of different bacterial taxons growing at high copper concentrations (340 - 512 μg/ml).
Children’s copper exposure and bacterial copper resistance
A substantial proportion of children’s fecal samples contains high copper-resistant (64%) or intermediate-copper resistant Gamma-Proteobacteria (95%). Resistance was significantly higher among organisms with environmental niches than in Escherichia coli, suggesting that environmental contamination with copper might be at the origin of copper-resistant bacteria of the children’s gut. A high proportion of children’s fecal samples (90%) contained copper-resistant Firmicutes, with high predominance of Enterococcus, particularly E. faecium.
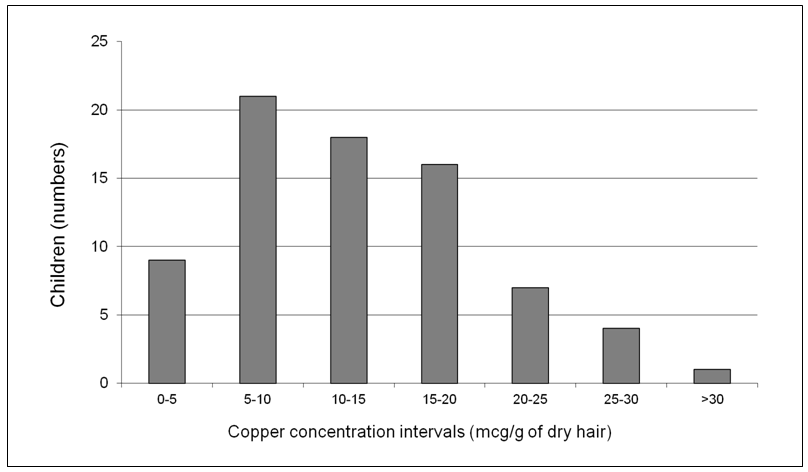
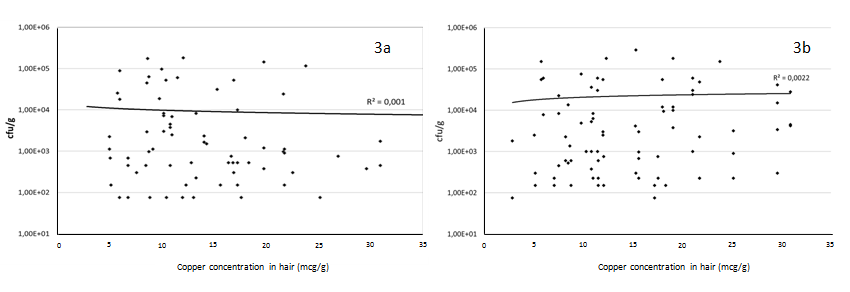
Our group of healthy children had various degrees of copper-concentration in the hair samples, ranging from ≤ 5 to more than 30 mcg/g in dry hair (Figure 2). In order to determine the degree of relationship between the microorganism survival found in a sample and copper levels measured in the sample, the density (colony counts/ml) of the bacteria belonging to the Gamma-Proteobacteria and Firmicutes taxa was plotted against the copper levels measured in the hair sample of the corresponding child, and the Pearson correlation (r) was obtained (Figure 3). In all cases, Pearson r correlation was close to zero (r = 0.001 and 0.0002, respectively), indicating the low dependence grade between these variables. Hence, we did not detect any direct (or inverse) relationship between environmental exposure to copper and the microbial counts. As stated in a previous paragraph, copper- resistance is significantly higher in predominantly environmental microorganisms, suggesting that environmental pollution with copper could be the source of copper resistance in bacteria from children’s intestine.
Figure 2: Copper concentrations (mg/g of dry hair) in children.
Figure 3: Correlation of copper concentrations (mg/g of dry hear) with bacterial counts of intestinal isolates growing in high copper concentrations (340 - 512 μg/ml). 3a, Gamma-Proteobacteria (recovered in gradient plates of McConkey Agar); 3b, Firmicutes (recovered in gradient plates of M-Enterococcus medium)
Discussion
To our knowledge this study provides the most detailed analysis available of copper-resistant microorganisms in intestinal human samples, and particularly in children. Current knowledge about copper-susceptibility of bacterial species from intestinal samples is mainly confined to animals, and based on the screening of Minimal Inhibitory Concentrations (MICs), of a small number of bacterial isolates per sample. Seeding the samples in antimicrobial gradients (as Szybalsky plates) allows evaluation of the effect of toxic agents on natural bacterial populations, particularly when different mechanisms of adaptive tolerance and resistance, that can result in changes in gene expression and a wide phenotypic diversity, can co-exist as in the case of copper. In fact, recent studies show that copper is able to decelerate bacterial growth or even reach lethal concentrations at millimole concentration[16]. Such diversity cannot be captured only by testing individual colonies with conventional MIC methods. Some previous studies focused on particular bacterial groups have used steep gradient plates to detect the phenotype provided by certain genes[17], but never to detect copper-resistance in natural samples, which require a flatter gradient to differentiate small differences in susceptibility.
There are several mechanisms by which bacteria tolerates various levels of copper in their environment. Copper sensing and tolerance might result from relatively unspecific stress response genes[18]. Note that in most cases of copper resistance there is a fuzzy difference between tolerance and resistance, as there are mechanisms of “metalloregulation” in which different genetic mechanisms overlap their phenotypes to protect bacteria against a wide range of toxic (producing reduction of growth rate) copper concentrations, from low to high ones[19,20].
Mechanisms for copper resistance in Gamma Proteobacteria include efflux systems (CusCFBA, CopA-B), copper-binding (CusF, siderophores) and copper-oxidation (peroxidases, CueO). However, many “unspecific mechanisms”, such as multidrug efflux transporters (as AcrAB from E. coli, or MexB in P. aeruginosa), periplasmic chaperones, or proteins involved in the response to superoxydes, act to reduce copper toxicity[21]. Extrachromosomal elements might encode resistance mechanisms to high copper concentrations, frequently regulated by copper-dependent transcriptional regulatory systems (CueR, CusRS, PcoRS)[22]. In the phylum Firmicutes an efflux system, CopA-B, and the CueO gene oxidizing Cu(I) to the less toxic Cu(II), contributes to copper-resistance. A very effective mechanism (involving copper exporting ATPase), elicited by the tcrYAZB operon, assures resistance of exposure to high copper toxic concentrations (> 12 mM)[23-26]. Interestingly, CueO and tcrYAZB genes are closely linked, and might be co-regulated by copper sensors CucS, CusR); these genes are often located besides others that also confer resistance to other heavy metals as zinc, arsenic or mercury[24].
We stress that such diversity of mechanisms and phenotypes cannot be reliably captured only by testing individual colonies with conventional MIC methods. Most published studies are focused on copper-resistance in Firmicutes, and a concentration of 12 mM has been considered as the copper-resistant breakpoint of the enterococci[1,2,26]. Of course (see below) the fecal concentrations of copper are probably much lower than the concentration of this “in vitro” breakpoint. Most probably many bacterial organisms in a bacterial population that contain genes encoding adaptive mechanisms of copper resistance might be inhibited at much lower copper concentrations (intrapopulation phenotypic diversity). Consistently with this view, sub-inhibitory concentrations of heavy metals have been shown to be sufficient for selecting bacterial populations harboring multi-resistant plasmids with heavy-metal resistances[27]. Concentrations of copper around 1 mM have some effect on Gram-negative sewage bacteria[28]. In fact our study was designed to observe the effect of these relatively low copper concentrations (0 - 2.05 mM).
Which is the copper concentration that might influence the structure of bacterial populations in the intestine? This concentration should depend on the copper uptake. The amount of copper in feed used in growth promotion in pigs[29], frequently exceeds 50 times the amount required for normal growth, and a correlation was found between supplementation of pig feed with such a high copper exposure and the selection of Enterococci with specific mechanisms of resistance, as TcrB[30]. However, an increase of 10 fold the recommended concentration for growth does not affect the MICs of E. coli or Enterococcus recovered from feces[25], demonstrating again the difficulty of getting conclusions just from MIC values. It is possible that, as occurs insoil, exposure to low copper concentrations might strengthen the resistance of microorganisms to a subsequent copper stress[31]. If data on dose-response relation for Cu exposure and resistance are lacking, it seems likely that a resistance-driven effect in animals occurs at high trace element exposure rather than at more basal exposure levels[32].
In normal warm-blooded animals ingested copper is absorbed in the stomach and upper small intestine where the maximal concentrations (eventually reaching 10 mM) are expected to occur[19]. In humans, a copper is micronutrient obtained from food and drinking water, which contributes from 0.1 – 1 mg/day, with a tolerated maximum of about 5 mcg/ml[3]; 40% of dietary copper comes from food[3]. Interestingly, one of the key factors of children’s increased exposure to copper under natural conditions is the contamination with soil in the environment[33].
Intestinal copper absorption occurs very efficiently, and copper excess is removed by the liver and excretion with bile into feces, in the order of 4.5 mg/day[6]. However, most of the copper secreted with the bile is reabsorbed by enterocytes[34,35], and the copper concentration in Chinese children’s blood is around 1 mcg/ml[36]. In addition, the intestinal copper absorption is inversely proportional to the copper content in the diet, being reduced to about one-tenth in high-copper diets[37]. Based on these studies and estimations performed in human samples from autopsies[38], we estimate that the colonic and fecal concentrations of copper would be in the range of 1-2 mcg/ml.
The range of copper concentrations found in dry hair in this study (10 - 30 mcg/g) corresponds to that found in other studies[39]. It is of note that the infants’ hair copper levels decline towards the end of the first year of age[40]. Our data on bacterial susceptibility indicate that the expected amount of copper in the children’s gut is probably insufficient for selection of bacterial variants with increased copper resistance. In fact, the better adapted Proteobacterial species to the gut, E. coli, is significantly less resistant to copper than other Proteobacteria which retain a significant set of functions related with environmental lifestyle. Klebsiella organisms contribute to biochemical (as nitrogen fixation) and geochemical earth processes, and have been identified as major components of the microflora in several types of stressed nonclinical environments, including plant surfaces[41,42]. Similar features apply to some of the organisms more frequently interacting with the free environment, as Citrobacter, or Enterobacter. In Firmicutes, and in the case of Enterococcus, even if E. faecalis is more frequent than E. faecium in the intestine of humans, both species are able to survive in the environment and be exposed to copper[43].
Our proposal is that most bacteria with increased copper-resistance recovered from the children’s gut have evolved in the environment, and not in the human intestine. Probably not in tap water, where the maximum concentration (5 mcg/ml) is about 50 - 100 times lower than the concentrations used in this work to detect copper-resistant bacteria.
The concentration of copper in the environment is obviously variable[44], but concentrations of 10 - 60 mcg/g (roughly/ml) have been found in dry European agricultural soils[45]. Highly efficient resistance mechanisms might have been selected at places where copper concentration exceeds 300 mcg/ml, and these concentration peaks probably occur only in particular environmental areas. In fact, copper was detected in sediments of urban cities (as Stockholm) at a concentration as high as a 475 mcg/ml (Swedish Environmental Protection Agency), similar to the high ones used in our experiments of selection[46]. Usually water-soluble copper compounds occur in the environment after release through activities related with agriculture, mining, smelting, refining and coal-burning industries, and also by natural reasons, as weathering of or the solution of copper minerals[47,48]. Copper is also increasingly used as disinfectant, fungicide or algicide in wood preservation, engraving, lithography or petroleum refining[19]. Moreover, copper discharged to wastewater is concentrated during treatment[3]. In general, the concentration of copper in soil influences the evolution of copper-resistance in the environment at large[49]. Evolved copper-resistant bacteria from the environment might enter in the human intestine.
Our study indicates that among Gamma Proteobacteria E. coli, a specialist-intestinal adapted organism, has the lowest proportion of high copper-resistant isolates; however, it is still the most frequent copper-resistant organism in human samples, even in quantitative terms (high population size in the intestine). Other Gamma-Proteobacteria with less intestinal specialization, and harboring traits of environmental lifestyle, seem to have a higher proportion of high-copper resistant isolates, but they are quantitatively much less represented, and their population size in the intestine is lower.
Among aerobic Firmicutes, high-copper resistant Enterococcus is very frequently copper-resistant (in particular E. faecium), occurs with high prevalence in intestinal samples and has high population sizes, so that its contribution to copper-resistance is the more relevant. Our results are indeed consistent with the observation that intakes of copper were positively associated with the rise of Firmicutes[3]. It should be noticed that the presence of Staphylococcus in the stool might be the result of perianal skin contamination, and does not necessarily represent intestinal colonization. It should be finally noticed that copper hair concentrations might not reflect blood concentrations[50], but they allow an easy, non-invasive way of measuring exposure and ingestion influencing intestinal microbiota (not necessarily systemic toxicity).
In summary, we have been unable to document any significant correlation between the copper concentrations in the hair of children and the concentration of copper-resistant Gamma Proteobacteria or Firmicutes in the intestinal samples, and we suggest that the origin of copper-resistance in the children’s intestine depends on the populations selected by copper in the external environment.
Conclusion
Acknowledge:
We thank the Instituto de Salud Carlos III for the support of ASV (Sara Borrel fellowship). We also acknowledge the European Development Regional Fund “A way to achieve Europe (ERDF) for cofounding the Plan Estatal de I+D+I 2013-2016 (PI12-01581) and CIBER (CB06/02/0053) actions.
Funding Information:
This work was supported by a Grant of the Ministry of Economy and Competitiveness of Spain, FIS (PI10/02588) to FB. This study was funded in part by grants from the European Union (FP7-ENV-2011 cod 282957 and HEALTH.2010.2.4.5-1), and from Spain: Instituto de Salud Carlos III and The Ministry of Health (Red INMA G03/176, CB06/02/0041, FIS- PI12/01890, FIS-PI041436, FIS- PI081151, FIS-PI06/0867, FIS-PS09/00090, FIS-FEDER: 03/1615, 04/1509, 04/1112, 04/1931, 05/1079, 05/1052, 06/1213, 07/0314, 09/02647, 11/01007, 11/02591, 11/02038, 13/1944, 13/2032, 14/00891, 14/01687, and Miguel Servet-FEDER CP11/0178 and CP15/025), the Conselleria de Sanitat, Generalitat Valenciana, Department of Health of the Basque Government (2005111093 and 2009111069), the Provincial Government of Gipuzkoa (DFG06/004 and DFG08/001) and the Generalitat de Catalunya-CIRIT (1999SGR 00241).
Competing financial interests:
The authors declare they have no actual or potential competing financial interests.
References
- 1. de Romana, D.L., Olivares, M., Uauy, R., et al. Risks and benefits of copper in light of new insights of copper homeostasis. (2011) J Trace Elem Med Biol 25(1): 3-13.
Pubmed || Crossref - 2. Carrothers, J.M., York, M.A., Brooker, S.L., et al. Fecal microbial community structure is Stable over time and related to variation in macronutrient and micronutrient Intakes in Lactating Women. (2015) J Nutr 145(10): 2379-2388.
Pubmed || Crossref - 3. WHO/SDE/WSH/03.04/88. Copper in Drinking-water. Background document for development of WHO Guidelines for Drinking-water Quality. (2004).
Crossref - 4. Georgopoulos, P.G., Roy, A., Yonone-Lioy, M.J., et al. Environmental copper: its dynamics and human exposure issues. (2001) J Toxicol Environ Health B Crit Rev 4(4): 341-394.
Pubmed || Crossref - 5. Cartwright, G.E, Wintrobe, M.M. Copper metabolism in normal subjects. (1964) Am J Clin Nutr 14(4): 224-232.
Pubmed || Crossref - 6. Turnlund, J.R. Human whole-body copper metabolism. (1998) Am J Clin Nutr 67(5 Suppl): 960S–964S.
Pubmed || Crossref - 7. Ysart, G., Miller, P., Croasdale, M., et al. 1997 UK Total Diet Study--dietary exposures to aluminium, arsenic, cadmium, chromium, copper, lead, mercury, nickel, selenium, tin and zinc. (2000) Food Addit Contam 17(9): 775-786.
Pubmed || Crossref - 8. Højberg, O., Canibe, N., Poulsen, H.D., et al. Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets. (2005) Appl Environ Microbiol 71(5): 2267-2277.
Pubmed || Crossref - 9. Pang, Y., Patterson, J.A., Applegate, T.J. The influence of copper concentration and source on ileal microbiota. (2009) Poult Sci 88(3): 586-592.
Pubmed || Crossref - 10. Fuller, R., Newland, L.G.M., Briggs, C.A.E., et al. The normal intestinal flora of the pig. IV. The effect of dietary supplements of penicillin, chlortetracycline or copper sulphate on the faecal flora. (1960) J Appl Bacteriol 23(2): 195-205.
Crossref - 11. Freitas, A.R., Coque, T.M., Novais, C., et al. Human and swine hosts share vancomycin-resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. (2011) J Clin Microbiol 49(3): 925-931.
Pubmed || Crossref - 12. Hobman JL, Crossman LC. Bacterial antimicrobial metal ion resistance. (2015) J Med Microbiol 64(Pt 5): 471-497.
Pubmed || Crossref - 13. Dusko Ehrlich, S., Meta HIT Consortium. Metagenomics of the intestinal microbiota: potential applications. (2010) Gastroenterol Clin Biol 34(Suppl 1): S23-S28.
Pubmed || Crossref - 14. Freire, C., Ramos, R., Lopez-Espinosa, M.J., et al. Hair mercury levels, fish consumption, and cognitive development in preschool children from Granada, Spain. (2010) Environ Res 110(1): 96-104.
Pubmed || Crossref - 15. Valenzuela, A.S., Benomar, N., Abriouel, H., et al. Biocide and copper tolerance in enterococci from different sources. (2013) J Food Prot 76(10): 1806-1809.
Pubmed || Crossref - 16. Cheknev, S.B., Vostrova, E.I., Apresova, M.A., et al. Deceleration of bacterial growth in Staphylococcus aureus and Pseudomonas aeruginosa cultures in the presence of copper and zinc cations.(2015) Zh Mikrobiol Epidemiol Immunobiol 2: 9-17.
Pubmed - 17. Hasman, H., Aarestrup, F.M. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. (2002) Antimicrob Agents Chemother 46(5): 1410-1416.
Pubmed || Crossref - 18. Hao, Z., Lou, H., Zhu, R., et al. The multiple antibiotic resistance regulators MarR is a copper sensor in Escherichia coli. (2014) Nat Chem Biol 10(1): 21-28.
Pubmed || Crossref - 19. Rensing, C., Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. (2003) FEMS Microbiol Rev 27(2-3): 197-213.
Pubmed || Crossref - 20. Berg, J., Brandt, K.K., Al-Soud, W.A., et al. Selection for Cu-tolerant bacterial communities with altered composition, but unaltered richness, via long-term Cu exposure. (2012) Appl Environ Microbiol 78(20): 7438–7446.
Pubmed || Crossref - 21. Teitzel, G.M., Geddie, A., De Long, S.K., et al. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. (2006) J Bacteriol 188(20): 7242-7256.
Pubmed || Crossref - 22. Grass, G., Rensing, C. Genes Involved in Copper Homeostasis in Escherichia coli. (2001) J Bacteriol 183(6): 2145–2147.
Pubmed || Crossref - 23. Solioz, M., Abicht, H.K., Mermod, M., et al. Response of Gram-positive bacteria to copper stress. (2010) J Biol Inorg Chem 15(1): 3–14.
Pubmed || Crossref - 24. Zhang, S., Wang, D., Wang, Y., et al. Genome sequences of copper resistant and sensitive Enterococcus faecalis strains isolated from copper-fed pigs in Denmark. (2015) Stand Genomic Sci 10:5.
Pubmed || Crossref - 25. Jacob, M.E., Fox, J.T., Nagaraja, T., et al. Effects of feeding elevated concentrations of copper and zinc on the antimicrobial susceptibilities of fecal bacteria in feed lot cattle. (2010) Foodborne Pathog Dis 7(6): 643-648.
Pubmed || Crossref - 26. Silveira, E., Freitas, A.R., Antunes, P., et al. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. (2014) J Antimicrob Chemother 69(4): 899-906.
Pubmed || Crossref - 27. Gullberg, E., Albrecht, L.M., Karlsson, C., et al. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. (2014) MBio 5(5): e01918-14.
Pubmed || Crossref - 28. Becerra-Castro, C., Machado, R.A., Vaz-Moreira, I., et al. Assessment of copper and zinc salts as selectors of antibiotic resistance in Gram-negative bacteria. (2015) Sci Total Environ 530-531: 367-372.
Pubmed || Crossref - 29. Apgar, G.A., Kornegay, E.T., Lindemann, M.D., et al. Evaluation of copper sulfate and a copper lysine complex as growth promoters for weanling swine. (1995) J Anim Sci 73(9): 2640–2646.
Pubmed || Crossref - 30. Amachawadi, R.G., Shelton, N.W., Shi, X., et al. Selection of fecal enterococci exhibiting tcrB-mediated copper resistance in pigs fed diets supplemented with copper. (2011) Appl Environ Microbiol 77(16): 5597-5603.
Pubmed || Crossref - 31. Li, J., Zheng, Y.M., Liu, Y.R., et al. Initial copper stress strengthens the resistance of soil microorganisms to a subsequent copper stress. (2014) Microb Ecol 67(4): 931-941.
Pubmed || Crossref - 32. Yazdankhah, S., Rudi, K., Bernhoft, A. Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. (2014) Microb Ecol Health Dis 26: 25.
Pubmed || Crossref - 33. Oyoo-Okoth, E., Admiraal, W., Osano, O., et al. Element profiles in hair and nails of children reflect the uptake from food and the environment. (2012) Environ Toxicol Chem 31(7): 1461-1469.
Pubmed || Crossref - 34. Linder, M.C., Hazegh-Azam, M. Copper biochemistry and molecular biology. (1996) Am J Clin Nutr 63(5): 797S–811S.
Pubmed || Crossref - 35. Zimnicka, A.M., Ivy, K., Kaplan, J.H. Acquisition of dietary copper: a role for anion transporters in intestinal apical copper uptake. (2011) Am J Physiol Cell Physiol 300(3): C588-C599.
Pubmed || Crossref - 36. Cao, J., Gao, Z., Yan, J., et al. Evaluation of trace elements and their relationship with growth and development of young children. (2016) Biol Trace Elem Res 171(2): 270-274.
Pubmed || Crossref - 37. Lönnerdal, B. Intestinal regulation of copper homeostasis: a developmental perspective. (2008) Am J Clin Nutr 88(3): 846S-850S.
Pubmed || Crossref - 38. Lech, T., Sadlik, J.K. Copper concentration in body tissues and fluids in normal subjects of southern Poland. (2007) Biol Trace Elem Res 118(1): 10-15.
Pubmed || Crossref - 39. McKenzie, J.M. Alteration of the zinc and copper concentration of hair. (1978) Am J Clin Nutr 31(3): 470-476.
Pubmed || Crossref - 40. Özden, T.A., Gökçay, G., Cantez, M.S., et al. Copper, zinc and iron levels in infants and their mothers during the first year of life: a prospective study. (2015) BMC Pediatr 15: 157.
Pubmed || Crossref - 41. Brown, C., Seidler, R.J. Potential pathogens in the environment: Klebsiella pneumoniae, a taxonomic and ecological enigma. (1973) Appl Microbiol 25(6): 900-904.
Pubmed || Crossref - 42. Brisse, S., Grimont, F., Grimont, P.A.D. The genus Klebsiella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, eds. The Prokaryotes. A Handbook on the Biology of Bacteria. 2006; 3rd edition ed. New York: Springer.
Crossref - 43. Fisher, K., Phillips, C. The ecology, epidemiology and virulence of Enterococcus. (2009) Microbiology 2009 155(Pt 6): 1749-1757.
Pubmed || Crossref - 44. Flemming, C.A., Trevors, J.T. Copper toxicity and chemistry in the environment: a review. (1989) Water, Air, and Soil Pollution 44(1): 143-158.
Crossref - 45. Heijerick, D.G., Van Sprang, P.A., Van Hyfte, A.D. Ambient copper concentrations in agricultural and natural European soils: an overview. (2006) Environ Toxicol Chem 25(3): 858-864.
Pubmed || Crossref - 46. Swedish EPA. Environmental quality criteria: Lakes and Water courses. Report 5050 (2000) Stockholm Sweden.
Crossref - 47. Shirivastara, A.K. A review of copper pollution and its removal from water bodies by pollution control technology. (2009) Int J Environ Pollut 29(6): 552-560.
Crossref - 48. Martínez, C.E., Motto, H.L. Solubility of lead, zinc, and copper added to mineral soils. (2000) Env Pollu 107(1): 153-158.
Pubmed || Crossref - 49. Shi, Z., Cao, Z., Qin, D., et al. Correlation models between environmental factors and bacterial resistance to antimony and copper. (2013) PLoS One 8(10): e78533.
Pubmed || Crossref - 50. Rodrigues, J.L., Batista, B.L., Nunes, J.A., et al. Evaluation of the use of human hair for biomonitoring the deficiency of essential and exposure to toxic elements. (2008) Sci Total Environ 405(1-3): 370-376.
Pubmed || Crossref