Risk-Profile-for-Amyotrophic-Lateral-Sclerosis-(ALS) An-Approach-with-the-Study-on-Cytokine-Polymorphisms
Cristiana Pistono1, Chiara Boiocchi2, Stella Gagliardi3, Elena Alvisi4,5, Cristina Cereda3, Mariaclara Cuccia1*
Affiliation
- 1Laboratory of Immunogenetics, Department of Biology & Biotechnology L. Spallanzani, University of Pavia, Pavia, Italy
- 2Inter-department Multiple Sclerosis Research Centre, C. Mondino National Institute of Neurology Foundation, IRCCS, Pavia, Italy
- 3Laboratory of Experimental Neurobiology, IRCCS National Neurological Institute C. Mondino, Pavia, Italy
- 4Department of Neurological Sciences, University of Pavia, Pavia, Italy
- 5Division of General Neurology, IRCCS National Neurological Institute C. Mondino, Pavia, Italy
Corresponding Author
Mariaclara Cuccia, Head of Immunogenetics laboratory, Department of Biology and Biotechnology, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy, Tel: (39)0382-985529; Fax: (39)0382-528496; E-mail: mariaclara.cuccia@unipv.it
Citation
Cuccia, M., et al. Risk Profile for Amyotrophic Lateral Sclerosis (ALS): An Approach with the Study on Cytokine Polymorphisms. (2016) Int J Neurol Brain Disord 3(1): 1-3.
Copy rights
© 2016 Cuccia, M. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Amyotrophic Lateral Sclerosis; Cytokines Polymorphisms
Abstract
Amyotrophic Lateral Sclerosis (ALS) is a progressive neurodegenerative disorder where neuroinflammation plays an important role; previous data suggest that cytokines are strongly involved.
A group of ALS patients were studied for immune protein polymorphisms: a case-control study to determine statistically significant differences in allelic, genotypic and haplotypic frequencies of 22 genetic polymorphisms of 13 cytokines was performed. The aim was the detection of a risk profile for the potential identification of individuals prone to disease and the discovery of possible targets for medication or lifestyle modification. From statistical analysis a significant difference was observed in polymorphisms of IL-1β +3962 (genotype: CC p = 0.0347; CT p = 0.0149), TGF-β codon 25 (genotype: CG p = 0.0126; GG p = 0.013) and TNF-α -238 (genotype: AA p = 0.019). We take into consideration all possible haplotypes, observing a significance of TG haplotype in two IL-2 gene polymorphisms (-330, + 166; p =0.0038), and CC haplotype of TGF-β (codon 10, codon 25; p = 0.022).Furthermore, significant differences in IL-β1 and TNF-α mRNA expression between patients and controls were demonstrated. This is a starting study to understand the multiple factors involved in the onset and in the progression of ALS, with particular attention given on inflammatory genes.
Introduction
Amyotrophic lateral sclerosis (ALS) is a rare adult-onset motor neuron disease. Sporadic (SALS) and familial (FALS) forms are clinically and pathologically similar, suggesting a common pathogenesis[1]. However, the precise cause of this disease is still unknown. It is most likely that at least sporadic ALS is multifactorial. Many data support an immune system involvement[2]. Several studies showed increased number of circulating lymphocytes, increased expression of MHC class II molecules on monocytes, and higher level of inflammatory chemokines and cytokines, both in blood and in CNS[3,4].
Association between cytokine gene polymorphisms and circulating cytokines levels have been studied in many diseases, including neurodegenerative disorders[5,6]. We dedicated our attention to peculiar polymorphisms of several immune proteins, cytokines and their receptors to define differences in allelic and genotypic frequencies in patients and controls and possible variations in mRNA levels. We chose these particular polymorphisms because our interest in neuroinflammation. The aim of this investigation is the detection of a risk profile that will potentially allow an early identification of individuals susceptible to disease, in order to find potential targets for medication or possible lifestyle modification.
Materials and Methods
65 sporadic ALS patients, classified according to the El Escorial criteria[7], are enrolled for this study from Northern Italian population (34 were male and 31 female; the age of onset was 58.1 ± 3.2 months and the disease duration was 45.2 ± 4.8 months).
Controls, for allelic and genotypic cytokine polymorphisms, were obtained from data reported in the literature for the Italian population[8]. As control population for the haplotypic frequencies was used the Italian population data contained in “The Cytokine Anthropology Workshop” (13th International Histocompatibility Workshop, Seattle 2004). Blood cells from 35 age- and gender-matched healthy volunteers were used as controls for real-time, quantitative PCR experiments. The controls (healthy subjects free from pharmacological treatment and not affected by any infectious and inflammatory diseases as demonstrated by blood analysis (data not shown)) were recruited from Northern Italian population.
DNA of each individual was obtained from 1 ml of whole blood in EDTA using the GFX kit (GE Healthcare, Milan, Italy). For each patient a molecular study of 22 polymorphisms of 13 different cytokines and receptor proteins (IL-1α -889 T/C, IL-1β -511 C/T, IL-1β + 3962 T/C, IL-1R +1970 C/T, IL-1Rα + 11100 T/C, IL-4Rα +1902 G/A, IL-12 -1188C/A, IFN-γ+874 A/T, TGF-β1 codon 10 or +869 C/T, TGF-β1 codon 25 or +915 G/C, TNF-α -308 A/G, TNF-α-238 A/G, IL-2-330 T/G, IL-2 + 166 G/T, IL-4 -1098 T/G, IL-4 -590 T/C, IL-4 -33 T/C, IL-6 -174 G/C, IL-6nt565 G/A, IL-10 -1082 G/A, IL-10 -819 C/T, IL-10-592 C/A) was performed. All cytokine polymorphisms were investigated through SSP-PCR (Sequence Specific Primers- Polymerase Chain Reaction), using the Heidelberg kit (Cytokine Typing Tray kit, University of Heidelberg, Heidelberg, Germany).
Unambiguous haplotype reconstruction of cytokines polymorphisms has been made thanks to the building of primers, so that they could mutually recognize and form complementary bonds in flanking sequences of polymorphic genes (Cytokine Genotyping kit, Invitrogen, Milan, Italy).
Gene expression experiments were done using lymphocytes from a casual subgroup of 35 SALS patients. Total RNA was extracted with the Trizol® reagent (Invitrogen, Milan, Italy) and quantified by spectrophotometer analysis. One μg of RNA was reverse transcribed using iScriptcDNA Synthesis Kit (Bio-Rad, Segrate, Italy). TNF-α, IL-1β and TGF-β1 mRNA amounts have been analysed using Sybr-Green primers designed with identical annealing and melting temperatures using Primer Express Software (Applera, Milan, Italy). Real-Time PCR was done with iCycler PCR Detection System (iQ5, Bio-Rad, Segrate, Italy) using iQSupermix (Bio-Rad, Segrate, Italy).
Results and Discussion
By comparing the genotype of each polymorphism of ALS patients to healthy controls we observed a statistical significance in TGF-β1 codon 25 (CG genotype: p = 0.0126; GG genotype: p = 0.013), IL-1β +3962 (CC genotype: p = 0.0347; CT genotype: p = 0.0149), and TNF-β -238 (AA genotype: p = 0.019).
By comparing the allelic frequencies of each polymorphism of patients with the frequencies of controls we observed a significance only at codon 25 of TGF-β1 (p = 0.02).
From the unvaried analysis obtained comparing the haplotypes of each cytokine with the results of controls, we observed a significant difference of TG haplotype of IL-2 (-330, +166; p = 0.0038), and CC haplotype of TGF-β1 (codon 10, codon 25; p = 0.022) (Table 1).
Table 1: Haplotypic frequencies statistically significant in ALS patients and healthy controls.
| Haplotype | ALS | Controls (n = 88) | p | |
|---|---|---|---|---|
| IL-2 (-330; +160) | GT | 1% | 0% | 0,1894 |
| GG | 38% | 29% | 0,1118 | |
| TG | 28% | 46% | 0,0038 | |
| TT | 33% | 25% | 0,1867 | |
| TGF-β (cd10; cd25) | CC | 12% | 6% | 0,0228 |
| CG | 37% | 39% | 0,6093 | |
| TC | 2% | 0% | 0,1924 | |
| TG | 49% | 55% | 0,3251 |
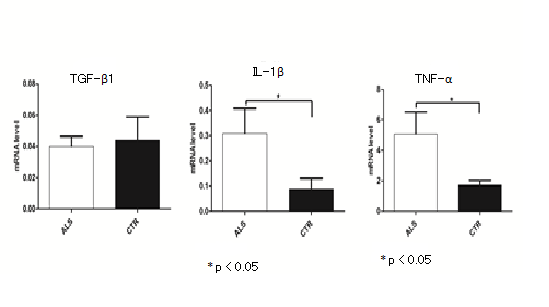
We also compared mRNA TGF-β1, TNF-α and IL-1β genes expression between ALS patients and controls. mRNAs of TNF-α and IL-1β were expressed at higher level in ALS patients than in controls (p < 0.05) (Figure 1). TGF-β1 mRNA quantity was similar in sporadic ALS samples and in controls. No correlation between genotypes and mRNA expression was found.
Figure 1: Expression of TGF-β1, IL-1β and TNF-α mRNA in lymphocytes of SALS patients. Healthy age- and gender-matched subjects were used as controls. The expression analysis was made using YWHAZ and UBC as housekeeping genes for normalization with 2-ΔΔCt comparative method. Student’s t-test showed a significantly different IL-1α and TNF-α expression (p < 0.05) between ALS patients and their respective controls. No differences in TGF-β1gene expression between patients and controls was observed.
A statistically significant increase in CC haplotype (codon 10, codon 25) of TGF-β1 (Table 1) was detected in patients. A statistically significant decrease in GG genotype and a statistically significant increase of CG genotype of TGF-β1 codon 25 polymorphism was observed in patients. TGF-β1 acts as an inhibitor of immune and inflammatory responses. TGF-β1 also performs several functions outside the immune system. Polymorphism G/C transversion at codon 25 of TGF-β1 gene generates an amino acid substitution from arginine to proline[9], that is supposed to have a role in controlling the concentration of TGF-β1[10]. However, from our data it seems that this polymorphism does not influence mRNA levels, according to TGF-β1 serum levels data, that were found increased only at the terminal status of disease[11].
The results of our study demonstrated a statistically significant increase of AA genotype of TNF-Α -238 polymorphism in promoter region of gene in ALS patients. TNF-α is the main mediator of the acute inflammatory response; it is also responsible for many events that can often complicates systemic infections. The most common G allele is associated with high production of TNF-α[12]. TNF-α high plasma concentration was present at disease onset and remains over the normal range during the whole disease progression time. We suppose that mRNA level increase may be due to an oxidative stress involvement in all patients analysed[13]. TNF-α could be a protective response to ROS:hydrogen peroxide is an activator of TNF-α gene expression through NF-κB[14]. The increase of TNF-α might be one of attempts of the cells to reduce SOD1 protein in patients[15].
About IL-1β the results suggest that the +3962 polymorphism could have an important role in susceptibility to disease; there was a statistically significant decrease of CT genotype of IL-1β +3962 polymorphism. Analysis showed a statistically significant increase of the CC genotype, not associated with an increased expression of this cytokine. Our data show a higher mRNA IL-1β expression in ALS patients than in controls. The relationship between ALS and IL-1β is supported by data in PC12 cells that demonstrated SOD1 down-regulation linked to IL-1β increase[16]. Furthermore, microglia in spinal cord of mSOD1 mice shows increased levels of NOX2 and IL-1β[17]. Oxidant stress condition, known in ALS disease, is a specific and important inducer of IL-1β expression[18].
Conclusion
This is a preliminary approach to the investigation on multiple factors involved in onset and progression of ALS, with particular attention given to the polymorphisms of genes for inflammatory molecules and to their expression. The demonstration of SOD1 mRNA increase in pathological nervous areas and in lymphocytes from ALS patients[19], and the evidence of the interaction between TNF-α, IL-1β and SOD[16], underline the specific role of cytokines in ALS. TNF-α and IL-1β over expression could have a protective function by means of down-regulation of SOD1.
Genetic cannot explain by itself the onset and the progression of this complex disease: we have to consider, together with unknown environmental factors, the role of epigenetic mechanisms. Epigenetic alterations have been observed in ALS patients[20] and can be important in driven the alteration of the immune system. Certainly, starting from these considerations, we plan to deeply investigate the relationship between TNF-α, IL-1β and SOD1 in this pathological state.
References
- 1. Gruzman, A., Wood, W.L., Alpert, E., et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. (2007) Proc Natl Acad Sci U S A 104(30):12524-12529.
- 2. Zhao, W., Beers, D.R., Appel, S.H. Immune-mediated mechanisms in the pathoprogression of amyotrophic lateral sclerosis. (2013) J Neuroimmune Pharmacol 8(4):888-899.
- 3. McCombe, P.A., Henderson, R.D. The Role of immune and inflammatory mechanisms in ALS. (2011) CurrMol Med 11(3):246-254.
- 4. Rentzos, M., Evangelopoulos, E., Sereti, E., et al. Alterations of T cell subsets in ALS: a systemic immune activation? (2012) Acta Neurol Scand 125(4):260-264.
- 5. Bidwell, J., Keen, L., Gallagher, G., et al. Cytokine gene polymorphism in human disease: on-line databases. (1999) Genes Immun 1(1):3-19.
- 6. Haukim, N., Bidwell, J.L., Smith, A.J., et al. Cytokine gene polymorphism in human disease: on-line databases, supplement 2. (2002) Genes Immun 3(6):313-330.
- 7. Brooks, B.R., Miller, R.G., Swash, M., et al. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. (2000) Amyotroph Lateral Scler Other Motor Neuron Disord 1(5):293-299.
- 8. Uboldi de Capei M.U., Dametto, E., Fasano, M.E., et al. Genotyping for cytokine polymorphisms: allele frequencies in the Italian population. (2003) Eur J Immunogenet 30(1):5-10.
- 9. Cambien, F., Ricard, S., Troesch, A., et al. Polymorphisms of the transforming growth factor-beta 1 gene in relation to myocardial infarction and blood pressure. The Etude Cas-Témoin de l'Infarctus du Myocarde (ECTIM) Study. (1996) Hypertension 28(5):881-887.
- 10. Stoll, C., Mengsteab, S., Stoll, D., et al. Analysis of polymorphic TGFB1 codons 10, 25, and 263 in a German patient group with non-syndromic cleft lip, alveolus, and palate compared with healthy adults. (2004) BMC Med Genet 5:15.
- 11. Ilzecka, J., Stelmasiak, Z., Dobosz, B. Transforming growth factor-Beta 1 (tgf-Beta 1) in patients with amyotrophic lateral sclerosis. (2002) Cytokine 20(5):239-243.
- 12. Wilson, A.G., Symons, J.A., McDowell, T.L., et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. (1997) Proc Natl Acad Sci U S A 4(7):3195-3199.
- 13. Barber, S.C., Shaw, P.J. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. (2010) Free Radic Biol Med 48(5):629-641.
- 14. Moodie, F.M., Marwick, J.A., Anderson, C.S., et al. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. (2004) FASEB J 18(15):1897-1899.
- 15. Cova, E., Cereda, C., Galli, A., et al. Modified expression of Bcl-2 and SOD1 proteins in lymphocytes from sporadic ALS patients. (2006) Neurosci Lett 399(3):186-190.
- 16. Fogal, B., Hewett, S.J. Interleukin-1beta: a bridge between inflammation and excitotoxicity? (2008) J Neurochem 106(1):1-23.
- 17. Beers, D.R., Zhao, W., Liao, B., et al. Neuroinflammation modulates distinct regional and temporal clinical responses in ALS mice. (2011) Brain Behav Immun 25(5):1025-1035.
- 18. Brabers, N.A., Nottet, H.S. Role of the pro-inflammatory cytokines TNF-alpha and IL-1beta in HIV-associated dementia. (2006) Eur J Clin Invest 36(7):447-458.
- 19. Gagliardi, S., Cova, E., Davin, A., et al. SOD1 mRNA expression in sporadic amyotrophic lateral sclerosis. (2010) Neurobiol Dis 39(2):198-203.
- 20. Paez-Colasante, X., Figueroa-Romero, C., Sakowski, S.A., et al. Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. (2015) Nat Rev Neurol 11(5):266-279.